Knowing Your NGS Downstream: Functional Predictions
Abstract
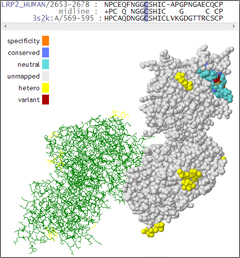
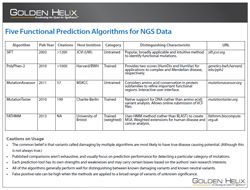
Next-Generation Sequencing analysis workflows typically lead to a list of candidate variants that may or may not be associated with the phenotype of interest. Any given analysis may result in tens, hundreds, or even thousands of genetic variants which must be screened and prioritized for experimental validation before a causal variant may be identified. To assist with this screening process, the field of bioinformatics has developed numerous algorithms to predict the functional consequences of genetic variants. Algorithms like SIFT and PolyPhen-2 are firmly established in the field and are cited frequently. Other tools, like MutationAssessor and FATHMM are newer and perhaps not known as well.
This presentation will review several of the functional prediction tools that are currently available to help researchers determine the functional consequences of genetic alterations. The biological principals underlying functional predictions will be discussed together with an overview of the methodology used by each of the predictive algorithms. Finally, we will discuss how these predictions can be accessed and used within the Golden Helix SNP & Variation Suite (SVS) software.
About the Presenter

Dr. Bryce Christensen fills two roles at Golden Helix as he is both the Director of Services as well as a Statistical Geneticist. Bryce joined GHI in 2009 from the University of Utah where he earned his PhD in Genetic Epidemiology and Biomedical Informatics. Before undertaking his graduate studies, Bryce worked for 2 years as a data analyst at Mayo Clinic in the Division of Biostatistics.