What’s Coming to VSClinical: Expanded Clinical Trial Search Results

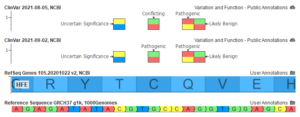
While the interpretation of germline variants generally focuses on the pathogenicity of a variant for a specific disease, the interpretation of somatic variants is centered around each variant’s impact on clinical care. As a result, clinical trials play an important role in assessing the clinical significance of somatic biomarkers, with…
Read more →