While VarSeq comes with a number of starter workflows that are stored as templates, customers also have the option of creating filter chains from scratch; analyzing a single exome may require you to do exactly that. In this blog, I’ll go through analyzing a single exome and generating a list of variants for further study. After importing the variant data… Read more »
The next release of VarSeq will ship a new product that is highly relevant to our customers in clinical testing labs. Via VSReports, VarSeq now has the ability to generate clinical-grade reports. These reports are fully customizable, containing focused and actionable data. VS Reports ships with report templates that are modeled off of the ACMG guidelines, the de-facto gold standard… Read more »
With our latest release of VarSeq last Thursday, we are proud to offer the VarSeq ® Viewer to the community, for free! When you download VarSeq ® Viewer, you can explore pre-built projects to interact with the annotations and filters the project provides, visualize selected variants with pile-ups, and export data to widely used formats. To get you started, we have included… Read more »
A widely attended conference that is coming up on September 21-23 is the third international conference on Genomics 2015 in San Antonio, Texas. The theme of this year’s conference is “Implications and Impacts of Genomic Advances on Global Health“, which promises to provide some interesting presentations and discussions. Current issues in the field to be addressed include next-gen sequencing, clinical and… Read more »
When it comes to down to it, the genomic variants we collect in a research and clinical setting are impossible to interpret without that important link of how genes are related to phenotypes. Indisputably, the Johns Hopkins project to catalog all evidence related to inheritable Mendelian diseases is our best repository of this evidence. Online Mendelian Inheritance in Man (OMIM)… Read more »
Have you ever scratched your head when looking up a variant and it seems like the number you have for its position is one off from what it looks like in the file or database? You may be running into the dreaded world of 1-based versus 0-based coordinate representation! If it’s any consolation, I can promise that all the bioinformaticians… Read more »
A prerequisite for clinical NGS interpretation is ensuring that the data being analyzed is of high enough quality to support the test results being returned to the physician. The keystone of this quality control process is coverage analysis. Coverage analysis has two distinct parts. Ensure that there is sufficient coverage to be confident in called variants Make certain that no… Read more »
I was definitely an early adopter when it comes to personal genomics. In a recent email to their customer base announcing their one millionth customer, they revealed that I was customer #44,299. And I have been consistently impressed with the product 23andMe provides through their web interface to make your hundreds of thousands of genotyped SNPs accessible and useful. It… Read more »
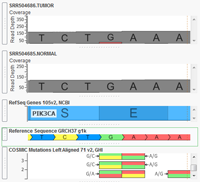
VarSeq now supports analysis of paired Tumor/Normal samples! Tumor/Normal support has been one of the most common feature requests for VarSeq since it was launched late last year, and we are excited to make this functionality available to all of our VarSeq users in the latest update (version 1.1.4). VarSeq is a powerful platform for annotation and filtering of DNA… Read more »
Recently, Golden Helix, Inc. announced the addition of VSPipeline to our VarSeq software suite. VSPipeline is a command-line interface that will allow high throughput environments the ability to tap the full power of VarSeq’s algorithms and flexible project template system from any command line context, including existing bioinformatics pipeline. So, what is the big deal? Here are the top five… Read more »
One of the lesser known functions in SVS allows the user to create Venn diagrams comparing variants found in multiple spreadsheets. These different spreadsheets could come from individuals samples, a case vs. control group or several variant databases. It is a helpful tool for visually comparing different variants. Start by creating spreadsheets for each group/samples you’d like to compare. The… Read more »
This last week I had the pleasure of attending the fourth annual Clinical Genome Conference (TCGC) in Japantown, San Francisco and kicking off the conference by teaching a short course on Personal Genomics Variant Analysis and Interpretation. Some highlights of the conference from my perspective: Talking about clinical genomics is no longer a wonder-fest of individual case studies, but a… Read more »
Just a few weeks ago we announced our partnership with MedGenome. The news was covered by a number of outlets including: Financial Express Pharmabiz Let me expIain the importance and impact of this announcement. Since Varseq was released, we have received strong interest from testing labs that are leveraging our product to implement cancer diagnostic pipelines. Please feel free to take… Read more »
The support team at Golden Helix is always on-hand to help with your SVS and VarSeq needs. We get some questions more often than others, and this blog will answer some of the most common questions we’ve been seeing lately regarding VarSeq. A common question we receive is if data can be filtered from a locally kept set of variants… Read more »
There’s a strong desire in the genetics community for a set of canonical transcripts. It’s a completely understandable and reasonable thing to want since it would simplify many aspects of analysis and especially the downstream communicating and reporting of variants. Unfortunately, biology isn’t so tidy as to provide a clear answer for which transcript is the important one. Consequently, there… Read more »
Our VarSeq as a Clinical Platform webcast last week highlighted some recent updates in VarSeq that support gene panel screenings and rare variant diagnostics. The webcast generated some good questions, and I wanted to share them with you. If the questions below spark new questions or need clarification, feel free to get in touch with us at [email protected]. Question: Should… Read more »
As VarSeq has been evaluated and chosen by more and more clinical labs, I have come to respect how unique each lab’s analytical use cases are. Different labs may specialize in cancer therapy management, specific hereditary disorders, focused gene panels or whole exomes. Some may expect to spend just minutes validating the analytics and the presence or absence of well-characterized… Read more »
Last week we conducted a webcast on “Cancer Gene Panels”. We had some excellent questions which we answered during the webcast and a few more that we didn’t get to in the allotted time. Please find answers to those questions here: 1. Are Cancer Gene Panels just another stepping stone on the way to whole exome/genome analysis? Cancer gene panels answer… Read more »
I am constantly on the lookout for fun or interesting datasets to analyze in SVS or VarSeq and recently came across a study looking into inherited cardiac conduction disease in an extended family (Lai et al. 2013). The researchers sequenced the exomes from five family members including three affected siblings and their unaffected mother and an unaffected child of one… Read more »
In just 5 days, the 22nd International Molecular Medicine Tri-Conference (Tri-Con) will kick off in San Francisco. This year, Tri-Con will offer over 3,000 attendees 6 symposia, over 20 short courses, and 17 conference programs focused on drug discovery, genomics, diagnostics, and information technology surrounding, molecular medicine. Both Dr. Andreas Scherer, CEO of Golden Helix and Gabe Rudy, our Vice President of… Read more »