This week we launched VSClincial with our first webcast to show our powerful new way to perform variant interpretation following the ACMG guidelines. Our audience asked a lot of great questions on the new product and I’d like to highlight a few here. Can VSClinical run on a laptop and/or a locked down environment? Like all of Golden Helix products, you have… Read more »
Relating human phenotypes to genotypes is the name of the game with OMIM, and as their website says, “is intended for use primarily by physicians and other professionals concerned with genetic disorders, by genetics researchers, and by advanced students in science and medicine.” The Online Mendelian Inheritance in Man (or OMIM) was originally created by Dr. Victor A. McKusick in… Read more »
There are many good reasons why the pursuit of the highest quality genomic interpretation would lead you to the latest human reference. It is more complete and fixes incorrect or partially missing genes that have known implications for human disease. While most major projects cataloging human populations have plans to re-do all their genomic alignments to the new human reference… Read more »
2017 was an incredibly prosperous year for Golden Helix; we released a handful of new features, announced new partnerships and completed our end-to-end architecture for clinical testing labs. Our webcast series has become a very popular way for our community to stay up-to-date with our new capabilities and best practices in genetic analysis using our software. We had three webcast… Read more »
The Golden Helix SNP & Variation Suite (SVS) platform is a powerful and versatile set of tools and algorithms for performing genomic research. That research spans from data originating on genotype micro-arrays to next-generation sequencing. While the majority of SVS users start with genotype data on their samples, any genomic information across a cohort can be used in our various… Read more »
First of all, I wish you a prosperous 2018 along with happiness and health for you and your loved ones. This next year comes with lots of anticipation. We at Golden Helix are looking forward to another year of growth and innovation. Over the last few years, we were able to build a large following of clients in the clinical space…. Read more »
We have a lot to thank the 1000 Genomes project for in the genomics community. By the collaborate efforts of many researchers and organizations, the project produced not only the first catalog of rare human variation but in the process standardized many things we take for granted, such as the VCF and BAM file formats. The variant frequencies of the… Read more »
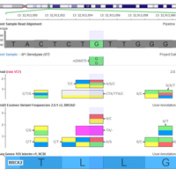
With the recent release of VarSeq 1.4.7, we have expanded the concepts of our popular assessment catalog to include CNV and other region-based records and not just variants. To match these capabilities, we have made a major update to VSWarehouse that supports these new record types in the centrally hosted and versioned Catalogs and Reports. Review of the VSWarehouse Genomic… Read more »
September 27, 2017 12:00 PM, EDT While Copy Number Variants are important to detect and interpret in many clinical genetic tests, labs have been without a comprehensive solution that integrates the annotating and reporting of high-quality CNV alongside their existing NGS variants. Golden Helix has developed and validated with our clinical partners a specialized NGS-based CNV caller capable of detecting… Read more »
An Example of an Integrated Clinical Workflow for CNVs and SNVs In this blog series, I discuss the architecture of a state of the art secondary pipeline that is able to detect single nucleotide variations (SNVs) and copy number variations (CNVs) in one test leveraging next-gen sequencing. In Part I, we reviewed genetic variation in humans and looked at the key… Read more »
The support team at Golden Helix is always here to help with your SVS and VarSeq needs. Often, we receive some excellent questions that should be shared with the rest of our users. This blog will answer some common questions we’ve been seeing lately regarding VarSeq CNV. I’ve noticed there is a version 2 of the CNV caller on Targeted Regions Algorithm, how has… Read more »
In case you missed our live event yesterday, I wanted to share the Q&A session and a link to the webcast recording: An Exploration of Clinical Workflows in VarSeq. Question:You mentioned saving projects as templates, will it save GenomeBrowse plots in the project template or do you have to replot the data when opening the software? Answer: Yes, if you save your… Read more »
We have been heads down doing the detailed and careful work to improve our CNV caller algorithm in the past three months since our we launched our Exome capable CNV caller and are very excited about the massive step forward we have made with the VarSeq 1.4.5 release. Additionally, we have added the all new Whole Genome large-event caller capable… Read more »
VarSeq enables breakthrough discoveries in cancer diagnostics by supporting gene panel testing and whole exome and genome analysis. We wanted to share our Cancer Gene Panel tutorial which covers a basic gene panel workflow with an emphasis on adding, modifying and manipulating filter chains. This tutorial will start with creating a new project from an empty project template, importing data, creating… Read more »
Annotating with gnomAD: Frequencies from 123,136 Exomes and 15,496 Genomes When the Broad Institute team lead by Dan MacArthur announced at ASHG 2016 that the successor to the popular ExAC project (frequencies of 61,486 exomes) was live at http://gnomad.broadinstitute.org/, I thought their servers would have a melt-down as everyone immediately jumped on and started looking up their favorite genes and… Read more »
The new VSWarehouse Tutorial covers the basic VSWarehouse workflow.This tutorial focuses on connecting to a VSWarehouse instance from VarSeq, adding an existing VSWarehouse project as an annotation source and using reports and assessment catalogs hosted on VSWarehouse. This workflow requires an active VarSeq license with the VSWarehouse feature included. You can go to Discover VarSeq or email [email protected] to request an… Read more »
In our latest VarSeq release, we updated our PhoRank algorithm with the ability to specify OMIM phenotype terms not present in HPO, as well as a general update to the algorithm to improve the results. In this post, we review the fundamentals of how PhoRank determines the ranking of genes in your VarSeq projects based on your input phenotype terms… Read more »
Recently, we added a natively supported Genotype Phasing and Imputation capability in SNP & Variation Suite 8.7.0. Since then we have had fantastic feedback and adoption as folks take advantage of the BEAGLE 4.0 and 4.1 algorithms from within their existing SNP GWAS and agrigenomic workflows. One piece of feedback we heard from our time at PAG, ACMG and our… Read more »
ACMG 2017 is just around the corner! We are halfway through March already and it’s just about time to head off to sunny and warm Phoenix, Arizona. While the temps have been mostly mild for the last few weeks in Montana, I bet those of you in the northeast are looking forward to your time in Phoenix! You will find… Read more »
Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS. This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes… Read more »