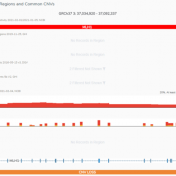
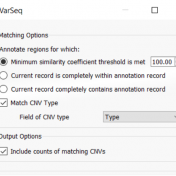
The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical. … Read more »
This morning I released a new version of my eBook “Clinical Variant Analysis – Second Edition.” The clinical interpretation of variants in Next-Gen Sequencing is a quickly evolving field. While the body of knowledge is growing exponentially, experts have to derive sound, clinical decisions leveraging an ever-expanding set of specialty databases, clinical publications, and algorithms that are designed to predict the… Read more »
Welcome to our Customer Publications for February 2021. As we commemorate the 57th American Heart Month, it is important to remember why we are wearing red and using #OurHearts. Heart disease is the leading cause of death worldwide. The National Heart, Lung, and Blood Institute urges Americans to do what they can to be heart-healthy and mitigate the risks of… Read more »
The recent release of VarSeq 2.2.2 brings our Word report template system, previously featured in VSClinical AMP, to the VSClinical ACMG workflow. This blog post will describe how to use the Word template system using one of our shipped templates as well as how to start customizing your own templates. We will cover the three different report templates that ship… Read more »
VarSeq recently received major upgrades in a wide range of areas, one of these areas includes adding annotations such as GnomAD. This includes new fundamental methods of CNV ACMG guideline processing but also a large number of small additions in annotations. One addition is the application of gnomADs – Gene Constraints. This provides various metrics for pathogenicity on a per… Read more »
New discoveries using NGS data analysis are never-ending and are pushing precision medicine to the forefront. In this month’s customer publication blog, I am focusing on our VarSeq software as investigators harness its power to perform a variety of investigational study designs. From cancer to inherited rare disease research, VarSeq is the rising star in research and diagnostic tools. At… Read more »
In continuation of our blog posts focusing on new features of VarSeq v2.2.2, here we will discuss the Latest Sample Assessment algorithm for both single nucleotide variants (SNVs) and copy number variants (CNVS). This algorithm annotates the variants of the project with the latest assessment from your variant catalog, which will show the history of interpretations made for the variants… Read more »
Our recent release of VarSeq 2.2.2 comes with a long list of upgrades and new features. In this blog post, we will demonstrate how defining sample phenotypes are available in VSClinical. One noticeable change is the ACMG guideline variant evaluation in VSClinical. Not only has this interface added CNV guideline evaluation, simplified the reporting process with embedded Microsoft Word and… Read more »
Golden Helix continues to innovate and grow, and due to this, we have been garnering attention and praise. We are thrilled to share with you some of the accolades and interest from numerous publications from the last six months. Talking points ranged from diagnosing and tracking Covid-19 infections leveraging NGS to new workflows, interpreting and reporting on copy number variants… Read more »
Happy New Year! I hope you had an opportunity to relax over the holidays with your family and friends. It’s time to start talking about our plans for the New Year, but first, please allow me to review a few highlights from 2020 that helped build a foundation for our future. Growth: Golden Helix was named to the 2020 Inc… Read more »
Have you seen us in The Journal of Precision Medicine? Last week, our team released VarSeq 2.2.2 loaded with a number of updates and VSClinical’s highly-anticipated ACMG-CNV Guideline workflow! We have spent the past several months sharing webcasts and blogs on this new capability. We are honored to also have this new solution recognized in The Journal of Precision Medicine… Read more »
Our upcoming release of VarSeq is one of the largest we’ve ever had with our software! It comes with an extensive list of polishes and new features like our recently mentioned ACMG CNV classifier and a redesigned reporting interface with updated templates. Additionally, this new release is also paired with some major upgrades to our list of new and supported… Read more »
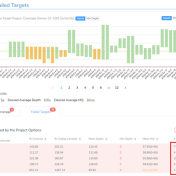
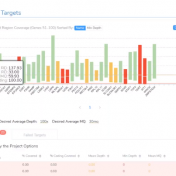
Curious about how coverage statistics can be used in conjunction with VarSeq? Evaluating the coverage over target regions or whole genomes is essential whether you are working with variant or CNV analysis. VarSeq has had the capability to compute sample level coverage statistics for some time now, but in the 2.2.2 release of VarSeq, there are some new features that… Read more »
Webcast Recap In the recent webcast “Exploring New Features and Clinical Reports in the ACMG Guideline Workflow”, Gabe and I took viewers through an evaluation with CNVs and SNVs according to the ACMG Guidelines where we generated and customized a clinical report. Along the way, we highlighted many new features that will soon be available in the upcoming VarSeq release…. Read more »
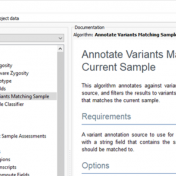
Those of you who have been attending our recent webcasts have learned about our upcoming VarSeq release. A part of that release will be an additional algorithm that will annotate variants matching the current sample. If you are not familiar with these webcasts, here are several on-demand webcasts I recommend to get you familiar with these new features: Evaluating Copy… Read more »
In lieu of conferences making the absolutely necessary decision to cancel this year’s conference, we created our Virtual 2020 Conference Booth! Even though it will never compare to the real deal, we hope you enjoy this blog post containing a few events we had planned this year. To start, let us paint the picture with some of our favorite conference… Read more »
As the world is consumed by the ongoing pandemic, it is easy to forget that there are investigators all around the globe that continue to make important discoveries in human medicine. Below are a few examples that remind us there are those that persevere in their chosen fields of study despite the trying times. At Golden Helix, we continue to… Read more »
In this blog update, I’ll be walking you through some of the advanced plotting capabilities with GenomeBrowse. The strategy with any next-gen sequencing analysis is to filter down to interesting variants for either research or clinical conclusion. Golden Helix produces powerful software specifically tailored for this efficient and comprehensive search for interesting and clinically relevant variants. One additional advantage of… Read more »
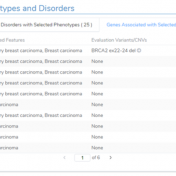
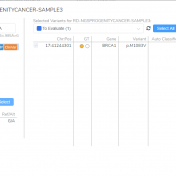
VSClinical is a feature to evaluate clinically relevant variants according to the ACMG or AMP guidelines. This feature can also be used to identify if a variant has been observed previously or evaluate a manually inserted variant. Take, for example, the scenario where a colleague is interested to see if you have seen any variants associated with Bechet syndrome, which… Read more »
We have now reached the final blog of the NGS-Solutions for Clinical Variant Analysis series. Part I of this series explored the capture of variant classifications in the VSClinical environment when following the ACMG and AMP guidelines. Part II was similar in content but for the capture of clinically relevant copy number variants as well as using a CNV catalog… Read more »