Golden Helix VSClinical provides a guided workflow interface for following the ACMG and AMP guidelines to evaluate variants and CNVs for NGS tests. The output of this work is most often a lab-specific clinical report. Since it was introduced, we have provided a powerful Word-based templating system to allow labs the ability to generate customized reports to include specific content… Read more »
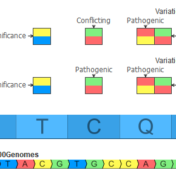
In the September 2021 monthly update to our curated ClinVar track, we made some changes that will result in roughly another 7,000 Likely Pathogenic and Pathogenic variants being available for annotation and use in the ACMG auto-classification system. Consensus Between Labs ClinVar has nearly one million unique variant classification records that are curated into multiple annotation tracks used in VarSeq and VSClinical on a monthly basis. Clinical… Read more »
I remember visiting a patient in the NICU amongst the incubators, some glowing blue like tiny tanning beds to treat jaundice, all containing tiny humans – many smaller than a loaf of bread. Infants get admitted into the neonatal intensive care unit or NICU for many reasons ranging from elevated bilirubin, hypoglycemia, sepsis, and respiratory distress (RDS). Many are eventually… Read more »
Next-gen sequencing (NGS) comprises many sophisticated steps that are often compressed into three major sections: library prep, sequencing, and data analysis. Obviously, the goal is to simplify each of these steps, but more often than not, there is a need for multiple tools to complete each one. Regarding the data analysis, Golden Helix seeks to provide simple yet comprehensive solutions… Read more »
VSPipeline is becoming a very popular tool among VarSeq users as it is essential for creating repeatable clinical workflows that can be executed in automated fashion. Since VSPipeline is a command-line tool, I think it would be helpful to discuss some of the best practices along with helpful tips for getting the most out of VSPipeline. Some of you may be less familiar with VSPipeline, so I want to cover how to set up the first run along with sharing the helpful tips as they arise. … Read more »
I learned about Batten disease from a childhood friend’s Facebook post. Over the course of a few months, her 8-year-old, Eva, the oldest of 4 daughters – Emily, Lucy, and Carly – was rapidly going blind. Baffled, doctors ran a genetic panel that returned a devastating result – the diagnosis of Juvenile Neuronal Ceroid Lipofuscinosis or Batten disease. A broad… Read more »
In many cases, VarSeq users design their next-gen sequencing workflows for a clinical application. One of the major values of using VarSeq is the standardization of sample analysis via project templates for filtering down to rare variants and isolate any clinically relevant variant. However, VarSeq also doubles as a robust research application as well. There are specific algorithms that can… Read more »
With the latest release of VarSeq, we have made significant updates to our handling of the interaction of variants and genes. This includes the support for non-coding transcripts, improved splice site predictions, and updates to gene and transcript annotations. We received several questions regarding how decisions are made in the software regarding genes and transcripts with these gene-related changes. This… Read more »
The GenomeAsia 100K Variant Frequency database is a pilot annotation source now available to our users. This valuable database offers a deep characterization of specific populations in Asia that can be used to drive genetic studies. GenomeAsia is comprised of whole-genome sequencing data of over 1,000 individuals from 219 populations across Asia. Using this as an annotation, users can analyze… Read more »
Thank you to those who attended the recent webcast, “VSWarehouse: Tracking Changing Variant Evidence and Classifications”. For those who could not attend but wish to watch, here is a link to the recording. The webcast covered some general highlights of VSWarehouse value but also presented some specific capabilities covering the ClinVar classification tracker. Golden Helix provides complete solutions to handle… Read more »
The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical. … Read more »
New discoveries using NGS data analysis are never-ending and are pushing precision medicine to the forefront. In the January 2021 customer publication blog, I am focusing on our VarSeq software as investigators harness its power to perform a variety of investigational study designs. From cancer to inherited rare disease research, VarSeq is the rising star in research and diagnostic tools…. Read more »
In continuation of our blog posts focusing on new features of VarSeq v2.2.2, here we will discuss the Latest Sample Assessment algorithm for both single nucleotide variants (SNVs) and copy number variants (CNVS). This algorithm annotates the variants of the project with the latest assessment from your variant catalog, which will show the history of interpretations made for the variants… Read more »
Webcast Recap In the recent webcast “Exploring New Features and Clinical Reports in the ACMG Guideline Workflow”, Gabe and I took viewers through an evaluation with CNVs and SNVs according to the ACMG Guidelines where we generated and customized a clinical report. Along the way, we highlighted many new features that will soon be available in the upcoming VarSeq release…. Read more »
In our previous webcast, Evaluating CNVs with VSClinical’s New ACMG Guidelines, we focused on a CNV deletion (12:27715515-29628122×1) in which the patient had a known disorder called Brachydactyly type E. The CNV was isolated using our VS-CNV caller and applied to the ACMG CNV guidelines using the intuitive steps of VSClinical. If you missed the webcast, you can watch the… Read more »
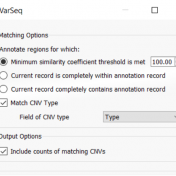
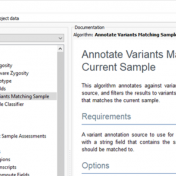
Those of you who have been attending our recent webcasts have learned about our upcoming VarSeq release. A part of that release will be an additional algorithm that will annotate variants matching the current sample. If you are not familiar with these webcasts, here are several on-demand webcasts I recommend to get you familiar with these new features: Evaluating Copy… Read more »
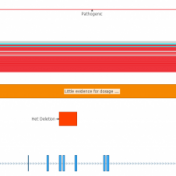
In the webcast, Evaluation of Copy Number Variants with VSClinical’s New ACMG Guideline Workflow, we discussed how VSClinical implements Section 4 of the ACMG guidelines. Specifically, we focused on integrating literature and publications to assess the pathogenicity of a CNV event when there was a lack of dosage sensitivity information. One of the primary pieces of evidence for evaluating genes… Read more »
In a recent webcast, users were exposed to some new features upcoming in the next release of VarSeq. In this update, we will use an example de Novo CNV in a cardiomyopathy panel in VarSeq. There is a long list of new tools and polishes to the software but one major upgrade is the inclusion of the ACMG and ClinGen… Read more »
A common discussion with our customers includes the challenges with the tertiary stage of analyzing next-gen sequencing data. This is the stage where all data from gene panels, exome, or whole genome scale pass through filters to quickly isolate the clinically relevant variant contributing to a patient disorder. Golden Helix has recognized these challenges in the scale of data and… Read more »
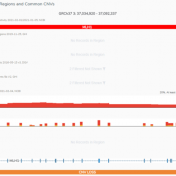
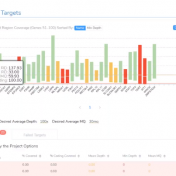
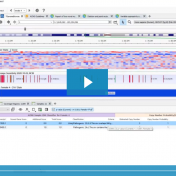
In this blog update, I’ll be walking you through some of the advanced plotting capabilities with GenomeBrowse. The strategy with any next-gen sequencing analysis is to filter down to interesting variants for either research or clinical conclusion. Golden Helix produces powerful software specifically tailored for this efficient and comprehensive search for interesting and clinically relevant variants. One additional advantage of… Read more »