We have a lot to thank the 1000 Genomes project for in the genomics community. By the collaborate efforts of many researchers and organizations, the project produced not only the first catalog of rare human variation but in the process standardized many things we take for granted, such as the VCF and BAM file formats. The variant frequencies of the… Read more »
Yesterday’s webcast, Genomic Prediction Methods in SVS, gave attendees a chance to see how the principles of genomic prediction are applied within SVS, predicting phenotypes for both plant and animal species. You can find a recording of the webcast on our site here should you be interested in checking it out or sharing with a colleague! The webcast garnered a… Read more »
It is a great honor for us to be recognized as one of the “Top 25 Biotechnology Solution Providers of 2017” by CIO Applications. You can find my interview with the editor here. We are incredibly thankful for the hundreds of organizations and thousands of users globally trusting our brand! Thank you for your support.
Golden Helix is excited to announce a new round of novel and updated annotations; including a frequency track, a region track, and a gene track. All three of these tracks were created with the use of VarSeq and its Convert Wizard functionality. First, the expansive 1000 genomes track (1kG) has been updated to include sub-population allele frequencies and heterozygous and… Read more »
With the recent release of VarSeq 1.4.7, we have expanded the concepts of our popular assessment catalog to include CNV and other region-based records and not just variants. To match these capabilities, we have made a major update to VSWarehouse that supports these new record types in the centrally hosted and versioned Catalogs and Reports. Review of the VSWarehouse Genomic… Read more »
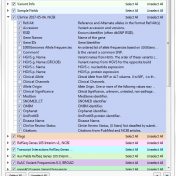
With the recent upgrade to VarSeq 1.4.7, users gain access to some new great features. Among the additions are new CNV annotations (Figure 1). In this final chapter of the annotation blog series, we are going to provide descriptions of the new CNV annotations and how they can be used. The types of CNV annotations vary and include frequency, clinical… Read more »
We’ve returned from another successful year at ASHG and had an incredible time. I find the conference to be a terrific opportunity to connect with our customers, partners, and friends in the industry. This year’s Presidential Symposium was a true delight featuring a discussion of global health and genomics between two absolute legends in the health and science world: Bill Gates,… Read more »
In our final chapter of this variant annotation blog series, we will discuss additional annotations that provide powerful variant filtering and analysis capability. Golden Helix curates many annotations in a way that allows for simple analysis and saves the users the hassle of all this data management. Whether you are trying to capture rare variants known across multiple subpopulations in… Read more »
VarSeq Updated with CNV Annotations, CNV PhoRank, and Region Assessment Catalogs This year we have released multiple advances to support CNV analysis in VarSeq, expanding our target region based VS-CNV caller to handle exomes and low-depth genomes as well as additional supporting algorithms like calling Loss of Heterozygosity regions. To top this off, VarSeq 1.4.7 has been shipped with many… Read more »
We look forward to seeing you all at ASHG 2017! Golden Helix is leaving the snowy mountains of Bozeman, MT and heading for the sunny streets of Orlando, FL to attend the 2017 American Society of Human Genetics Conference (ASHG)! We’re excited to be returning for the 12th year in a row and are looking forward to seeing everyone there…. Read more »
This month we hosted two, incredible webcasts officially announcing the latest CNV annotation capabilities our Software Engineering Team has been hard at work on for the past couple of months. Our first webcast, Comprehensive Clinical Workflows for Copy Number Variants in VarSeq, was presented by Golden Helix’s VP of Product & Engineering, Gabe Rudy, who reviewed the expanded capabilities of… Read more »
CIViC The Clinical Interpretations of Variants in Cancer, better known as CIViC, is an open access open source, community-driven web resource available to all VarSeq users. Nature Genetics published an article that states, “CIViC accepts public knowledge contributions but requires that experts review these submissions”. Fundamentally, the focus behind CIViC is to make sure the variants contained in the database… Read more »
Dr. Suzanne Lewis is a Clinical Professor in the Department of Medical Genetics at University of British Columbia (UBC) and Senior Investigator at British Columbia Children’s Hospital Research Institute (BCCHR), Vancouver, Canada. She is also the Chair of the iTARGET Autism Project and Vice-Chair of Autism Canada Chief Medical Officer and VP Research of Pacific Autism Family Network. She and… Read more »
Gabe Rudy gave a fantastic presentation yesterday on the latest additions to VS-CNV annotations. If you weren’t able to join us for the live event, you can access the recording and webcast slides here: Comprehensive Clinical Workflows for Copy Number Variants in VarSeq. Additionally, there were many great questions asked that we wanted to share with the community. Question: Can I… Read more »
Clinical Assessment Tracks Golden Helix provides a large catalog of annotation sources for our research and clinical clientele. Making these public data repositories available to all our users is no easy task. As Cody Sarrazin mentioned in his blog post, annotation curation is a complex data science pipeline. This process aggregates data from many disparate sources and normalizes it into… Read more »
September 27, 2017 12:00 PM, EDT While Copy Number Variants are important to detect and interpret in many clinical genetic tests, labs have been without a comprehensive solution that integrates the annotating and reporting of high-quality CNV alongside their existing NGS variants. Golden Helix has developed and validated with our clinical partners a specialized NGS-based CNV caller capable of detecting… Read more »
We find ourselves talking about our partnership with Sentieon, a lot! More specifically, we are always extolling the powerful, comprehensive genomic data analysis solution we are able to offer our clients. Sentieon’s Secondary Analysis Suite made a significant improvement in runtime over BWA-MEM, GATK, Mutect, and MuTect2 while providing deterministic and identical results. Here are two fantastic white papers that… Read more »
September 13, 2017 12:00 PM, EST This month we’re branching out and covering a topic we’ve never explored before – annotation capabilities! Darby Kammeraad, Field Application Scientist at Golden Helix, will be giving us some insight into the advantages of VarSeq’s capability with annotations. The number of annotation topics to cover are seemingly limitless. In this webcast, we will focus… Read more »
An Example of an Integrated Clinical Workflow for CNVs and SNVs In this blog series, I discuss the architecture of a state of the art secondary pipeline that is able to detect single nucleotide variations (SNVs) and copy number variations (CNVs) in one test leveraging next-gen sequencing. In Part I, we reviewed genetic variation in humans and looked at the key… Read more »
It’s fascinating to hear the various ways our products are being used by customers all around the world. This month we’re featuring publications citing VarSeq, SVS and HelixTree which cover studies from Schizophrenia, Multiple Sclerosis and more. We hope you enjoy! Exome sequencing in schizophrenic patients with high levels of homozygosity identifies novel and extremely rare mutations in the GABA/glutamatergic pathways This… Read more »