Join our upcoming webcast : Wednesday, April 5th 12:00 pm EST Dr. Reza Sailani is a Research Fellow in the Genetics department at Stanford University. To provide an overview of his research, Sailani will present on the following two recent studies he has conducted: Association of AHSG with alopecia and mental retardation (APMR) syndrome: Alopecia with mental retardation syndrome (APMR) is… Read more »
Today, I am happy to announce the introduction of VS-CNV for Gene Panels and Exomes. We have developed this capability in partnership with PreventionGenetics. PreventionGenetics will use VarSeq CNV for analysis of gene panels, and in the future for exome sequencing. The software gives PreventionGenetics the opportunity to conduct a comprehensive CNV analysis on NGS data, in many cases eliminating the need… Read more »
ACMG 2017 is just around the corner! We are halfway through March already and it’s just about time to head off to sunny and warm Phoenix, Arizona. While the temps have been mostly mild for the last few weeks in Montana, I bet those of you in the northeast are looking forward to your time in Phoenix! You will find… Read more »
Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS. This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes… Read more »
In the past couple of weeks, the topic of the Filter and Quality fields in the popular ExAC population catalog has come up a number of times. It turns out that unlike the 1000 Genomes project, which decided to very heavily filter their variant list to only contain variants they consider high quality, ExAC chose to include more dubious variants… Read more »
CIO Review recently published its annual Biotech Technology Special Edition announcing the 20 most promising biotech technology solution providers. It is a great honor that Golden Helix has been named among the top 20 for the second year in a row! Please find my interview with the editor here: Spearheading Innovation in the Biotech Industry We are very thankful for the hundreds of organizations and… Read more »
This year’s annual Abstract Challenge was once again a successful event. We want to thank all that participated. This year we had nearly 50 submissions and our decision was a difficult one. Here are the selected winners. This year, we had dual first place winners; Reza Sailani and Jingga Inlora of the Michael Snyder lab in the Department of Genetics at… Read more »
We are pleased to announce our next webcast, Calling Large LOH and CNV Events with NGS Exomes. The live event is scheduled for Wednesday, March 8th at noon EST. Here are the specifics: Wednesday, March 8th 12:00 pm EST Next Generation Sequencing Exomes are a powerful assay used in both clinical and research settings to discover novel and rare small variants…. Read more »
In the last year, Golden Helix has received several honors. We attribute these great achievements to our customers and the community. As we strive to provide the best analytics software available for both researchers and clinicians, the continued support, praise and referrals are monumental in our success. Pharma Tech Outlook selected Golden Helix as one of the top 10 Pharma… Read more »
This generation of scientists, clinicians and bioinformaticians have already elevated the standards for diagnosis, prediction and care, ultimately improving patient outcome for millions of people by leveraging genomic information. This trend is only going to continue. Next-gen sequencing has made its way into the clinic. Golden Helix supports the adoption of Precision Medicine by building products, such as our VarSeq… Read more »
In the new Genotype Imputation tool that is coming soon to SVS, allele encoding is an important part of matching data between the target and the reference panels. If the same platform provider is being used, then A/B encoding can be used. However, it’s better to use the Reference/Alternate allele encoding associated with AGCT format to ensure accuracy. If an… Read more »
Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results. Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs… Read more »
Genome-wide association study (GWAS) technology has been a primary method for identifying the genes responsible for diseases and other traits for the past ten years. GWAS continues to be highly relevant as a scientific method. Over 2000 human GWAS reports now appear in scientific journals. In fact, we see its adoption increasing beyond the human-centric research into the world of… Read more »
Join our upcoming webcast – Clinical Reporting Made Easy Wednesday, February 15th 12:00 pm EST Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the… Read more »
Just as we expected, 2017 has kicked off with a flurry of new publications by our customers. We even had a publication from a client using our VarSeq software! Congratulations to all, please take a look at some of the articles we have highlighted below: Reza Sailani of Stanford University and colleagues published Association of AHSG with alopecia and mental retardation… Read more »
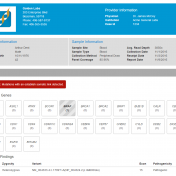
With a properly defined wet-lab and bioinformatics process, we are able to zero in on clinically relevant variants. How does a lab report on the outcome of their analysis? We find that most laboratories conduct their variant classification based on the guidelines formulated by the American College of Medical Genetics (ACMG) for inherited diseases. The ACMG guidelines for variant classification… Read more »
It may have been easy to miss in the drum-beat of monthly annotation updates we do here at Golden Helix, but there are a couple of things that are very special about the January update to the ClinVar database: We added new fields including HGVS names of variants and citations in PubMed for variants ClinVar nearly doubled in size by… Read more »
Acute lymphoblastic leukemia (ALL) is the most frequently diagnosed cancer in children and one of the leading causes of death due to disease in children. Dr. Daniel Sinnett, along with Pascal St-Onge and their colleagues at Sainte-Justine University Health center have been investigating the molecular determinants of the disease to improve detection, diagnosis and treatment. One particular area of study… Read more »
The year 2017 is starting fast and furious for us here at Golden Helix. We just announced a new imputation capability for our SVS product. At the same time, members of our team are on the way to PAG in San Diego to network with our clients in the Plant and Animal community. We have a terrific plan in place… Read more »
Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the results of tertiary analysis into a customizable clinical report. To include the VSReports functionality in… Read more »