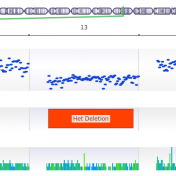
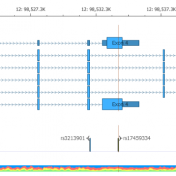
It is common knowledge that variants can be germline or somatic depending on whether the variant was inherited or acquired after birth. A well-known example is cancer-causing mutations in the BRCA genes, wherein the mutation may or may not have been inherited. Understanding the origin of the cancer-causing mutation is important when assessing potential treatment options as well as identifying… Read more »
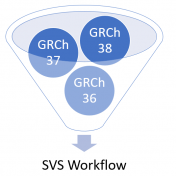
The world has been making a shift to use GRCh38 human genome reference coordinates, but the transition has not been fast. Many of the mainstay human catalog projects are changing to use native GRCh38 catalogs, or are remapping their current data to GRCh38 coordinates. While this seems to be the advancing goal, it is leaving researchers and analysts with the… Read more »
Thank you to everyone who joined our webcast, “Whole Genome Trait Association in SVS.” If you missed the live event and are interested in knowing what we talked about, you may access the recorded event below: Our Live Q&A generated a lot of great questions. Unfortunately, we were unable to answer them all, but we have compiled some of the… Read more »
Introduction: Malignant Rhabdoid tumors (MRT) are among the most aggressive and lethal forms of infant and child cancer (1). These tumors are characterized by an unusual combination of mixed cellular elements similar to but not typical of teratomas and can originate at any anatomic location. When MRTs are present in the brain, they are called atypical teratoid/rhabdoid tumors (AT/RT), which… Read more »
It is an honor to be published in the European Journal of Clinical and Biomedical Sciences (Volume 6, Issue 4) in an article co-authored by Dr. Christiane Scherer and myself, “Investigating the Global Spread of SARS-CoV-2 Leveraging Next-Gen Sequencing and Principal Component Analysis.” This study highlights how genomic surveillance of COVID-19 using Next-Generation Sequencing (NGS) can uncover transmission patterns and mutation… Read more »
Golden Helix has been incredibly fortunate to have been featured in a variety of publications over these last six months. Topics span from the history and future of our company to several new use cases for our solutions that extend into the infectious disease space, recently coming to fruition with the COVID-19 pandemic. We are so grateful to have received… Read more »
As I prepared to write the Customer Publication blog for June 2020, I was excited by the number of recently published papers that stood as examples of how both VarSeq and SVS software are employed to advance diagnostics and treatments in human medicine. We often think of SVS as the go-to platform for Agrigenomics, however both of our platforms have… Read more »
It is an honor to be featured in the Clinical OMICs May/June 2020 issue in a Q&A with the Editor discussing the past, present, and future of Golden Helix. In this article, I detail: Clinical OMICs Article: What has been Golden Helix’s most significant success or contribution to the industry over the past five years? Golden Helix started in 1998 with a… Read more »
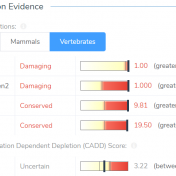
The University of Washington’s Combined Annotation Dependent Depletion (CADD) algorithm, now available in CADD v1.4 and CADD v1.5, measures the deleteriousness of genetic variants. This includes single nucleotide polymorphisms (SNVs) and short insertions and deletions (indels) throughout the human reference genome assembly. This algorithm was introduced in 2014 and has since become one of the most widely used tools to assess human… Read more »
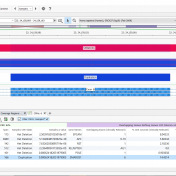
We have had many customers come to us over the years with a simple problem: they have BAM files for whole exome or gene panel data and would like to call CNVs using VarSeq’s powerful CNV calling capabilities, but they don’t have a bed file defining the target regions for their samples. To address this problem, we have developed a… Read more »
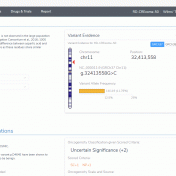
Abstract Before assessing the clinical significance of a somatic mutation, one must determine if the mutation is likely to be a driver mutation (i.e. a mutation that provides a selective growth advantage, thereby promoting cancer development). To aid clinicians in this process, VSClinical provides an oncogenicity scoring system, which uses a variety of metrics to classify a given somatic mutation… Read more »
An under-appreciated area of complexity when looking into the field of genetics from the outside can be found in genes and transcripts. Alternative splicing allows eukaryotic species to have a wonderfully powerful genetic code, resulting in multiple protein isoforms being encoded in a single section of DNA. But when it comes to variant interpretation, different transcripts can result in widely different predicted… Read more »
Like many of you, I was supposed to attend the 2020 ACMG Meeting in San Antonio this March with a few colleagues. In addition to attending the conference, my first blog post for Golden Helix was going to be about our time at the conference. Thanks to the world-wide COVID-19 pandemic, none of us got to attend the conference. Part… Read more »
Dear All, Going back in time to the beginning of this year, it would have been difficult to imagine what our world would look like as of June 2020. Things changed quickly. While we are still in the midst of the COVID-19 pandemic, the senseless killing of George Floyd triggered social unrest here in the USA that points to deeper… Read more »
Writing the blog post to summarize and highlight our customer’s publications is undoubtedly one of my favorite things to do! The wide variety of topics is always surprising and inspiring, and I am humbled by the efforts of dedicated scientists who are helping to protect and enrich our lives in so many ways. Our SNP & Variation Suite (SVS) software… Read more »
Today, we’re thrilled to announce that Healthcare Tech Outlook has named Golden Helix among the Top 10 Genetic Diagnostics Companies of 2020. This recognition places us alongside a remarkable group of companies that are leading innovation in genomics, diagnostics, and precision medicine. We are honored to receive this distinction and grateful for the opportunity to share our story in the… Read more »
In VarSeq 2.2.1, you can enable auto-updating for template annotation sources, ensuring they always reflect the latest available version. Previously, VarSeq templates were frozen in time. Now, each new project created from a template would use the same source that was used when the template was created. When you save a template, you can have the sources automatically update to… Read more »
SVS offers several options to conduct genome wide association tests and mixed linear models. At times, it can be challenging to decide which test, model, or adjustments to use when setting up your analysis. I want to briefly explore the options available in SVS for association tests, and mixed linear models to hopefully facilitate in understanding and choosing which options… Read more »
At Golden Helix, we’re committed to advancing precision oncology through robust software solutions that streamline clinical workflows. As the field of cancer genomics continues to evolve, staying informed about the latest best practices and tools is more important than ever. That’s why we’re excited to announce the release the second edition of Clinical Variant Analysis for Cancer eBook from our… Read more »
Thank you to everyone who joined me for our latest webcast, “Next-Gen Sequencing of the SARS-CoV-2 Virus with Golden Helix.” If you missed the live event and are interested in knowing what we talked about, you may access the recorded event below: Our Live Q&A generated a lot of great questions. Unfortunately, we were unable to answer them all, but… Read more »