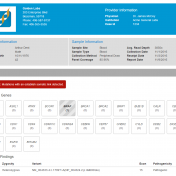
Generating a clinical report is the final step of most NGS pipelines and is important as it relays results and information to legacy systems, physicians and ultimately the patient. As reporting is a valuable process, Golden Helix offers reporting capabilities according to the ACMG and AMP guidelines but also as a standalone feature in VSReports. VSReports is a platform that… Read more »
The new VSReports tutorial covers a basic VSReports workflow with an emphasis on understanding and exploring report customizations. This tutorial requires an active VarSeq license with the the VSReport feature included. You can go to Discover VarSeq or email [email protected] to request an evaluation license with the VSReports functionality included. VS Reports provides the ability to generate clinical-grade reports. This feature enables VarSeq… Read more »
Join our upcoming webcast – Clinical Reporting Made Easy Wednesday, February 15th 12:00 pm EST Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the… Read more »
Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the results of tertiary analysis into a customizable clinical report. To include the VSReports functionality in… Read more »
Upcoming Golden Helix Webcast: Using Clinical Reports as part of a Gene Panel Pipeline Wednesday, July 13th @ 12:00 pm EDT VarSeq Reports can be used as part of an automatic pipeline to quickly list variants with information that can be used to make actionable clinical decisions in a readable HTML format. Need to further filter the variants or add interpretation… Read more »
Submit directly to N-of-One from VarSeq If you or your lab uses N-of-One solutions for clinical annotations, here’s some good news: You can now submit directly to N-of-One from VarSeq! N-of-One’s set of preferred transcripts may differ from those outputted by our algorithms in VarSeq, so our solution was built with that in mind. Our slick, easy to use, and… Read more »
Clinical reports come in all shapes, sizes and flavors. With that in mind, our clinical reporting interface VSReports was built to be highly customizable and flexible. With a little Javascript and HTML know-how, your clinical reports can be customized to meet the needs and goals of your lab. With a little Javascript and HTML know how, you can customize yours as… Read more »
The next release of VarSeq will ship a new product that is highly relevant to our customers in clinical testing labs. Via VSReports, VarSeq now has the ability to generate clinical-grade reports. These reports are fully customizable, containing focused and actionable data. VS Reports ships with report templates that are modeled off of the ACMG guidelines, the de-facto gold standard… Read more »
The week of October 10-16th was a busy time in our industry. Hundreds of biostatisticians, genetic epidemiologists, and statistical geneticists gathered in Cambridge, MA for the annual conference of the International Genetic Epidemiology Society (IGES) on October 10-12, followed by the biennial Genetic Analysis Workshop (GAW) on October 13-16. I had the opportunity to participate in both conferences, and I… Read more »
How much fun is a person allowed to have when going to a conference? That kind of sums up my personal experience going to the first ever Capita Selecta in Complex Disease Analysis (bi-annual) conference in Leuven, Belgium. Not only was it fun, but the conference program was packed with interesting talks and short courses. Dr. Kristel van Steen and… Read more »
As comprehensive genomic profiling (CGP) becomes the standard in cancer variant analysis, the scale and complexity of next-generation sequencing (NGS) data continue to grow. It is now commonplace to report on small variants, larger amplifications and deletions, various complex structural variants (fusions or breakends) as well as genomic signatures like microsatellite instability (MSI) and tumor mutation burden (TMB). Interpreting this… Read more »
Next-generation sequencing (NGS) data analysis often involves multiple steps from variant annotation, filtering, interpretation, visualization, and report generation. Repeating these steps manually for each new project can be error-prone and inefficient. In clinical and regulated environments, we recognize that consistency is critical. That’s why VarSeq offers robust support for workflow templates, enabling users to save, share, and reuse entire analysis… Read more »
In the evolving landscape of genomic medicine, accurate interpretation of complex genetic data is critical for uncovering the molecular underpinnings of rare and undiagnosed diseases. VarSeq, Golden Helix’s robust and intuitive variant analysis platform, continues to empower researchers and clinicians worldwide in this mission. Recent publications highlight how VarSeq enabled novel insights across diverse cases—from the identification of UGDH as… Read more »
The VarSeq platform has been on the market for over a decade and has established itself as robust and powerful clinical-grade bioinformatics software. Its usage spans many types of analysis, including pharmacogenomics, rare disease, hereditary cancer, somatic, carrier risk, and trio/family. Fundamentally, users have the freedom to build any workflow they need, which is a significant value proposition compared to… Read more »
ClinVar is a global, publicly accessible database maintained by the National Center for Biotechnology Information (NCBI) that archives interpretations of human genetic variants and their clinical relevance. ClinVar consists of contributions from clinical labs, expert panels, and research groups, and serves as a community-powered knowledge base that is foundational for clinical genetics. This powerful database is expectedly one of the… Read more »
VarSeq, when used with VSPipeline, automates carrier screening analysis, allowing labs to run them in a parallelized, economical fashion.
NGS Software is Bridging the Gap Between Discovery and Practical Application Researchers, clinicians, and scientists need cutting-edge solutions to better understand the complexities of human genetics. By delivering high-quality analysis tools and advancing the interpretation of genomic data, Golden Helix bridges the gap between discovery and practical application. Our NGS technologies play a critical role in accelerating genetic insights, supporting… Read more »
Thank you to everyone who joined our recent webcast, “Powering Genomic Workflows with Upgraded Catalogs in VSWarehouse and VarSeq 3,” presented by Gabe Rudy on April 23rd, 2025. We appreciate the engagement and interest in the latest advancements to our platform. For those who missed it or need a recap, the session focused on the pivotal role Catalogs play as… Read more »
Streamline rare and undiagnosed disease analysis with VarSeq—NGS software designed to simplify variant interpretation and diagnosis insights.
Genomic research is advancing at an unprecedented pace, and with it comes the need for powerful tools that can turn raw sequencing data into meaningful insights. VarSeq, our variant analysis and interpretation platform, has become a go-to solution for researchers tackling complex genetic questions. Whether in rare disease diagnostics, population genetics, cancer research, or infectious disease studies, VarSeq provides the… Read more »