In the first two parts of this blog, we presented examples of how to leverage Warehouse-stored VSClinical and CNV assessment catalogs in the VarSeq project. Now we are going to explore the Warehouse interface a bit more and show how to query on stored variant data. To access Warehouse from VarSeq, click the V Connect icon located in the top left corner of the project screen (Fig 1).

From the terminal (Fig 2) you will find multiple tabs:
Projects: Add new sample data to a previously stored project, edit preexisting projects/update templates, and create new projects to be uploaded to Warehouse
Reports: Use previously stored reports, modify, and upload new report into Warehouse
Catalogs: Open, modify and create new assessment catalogs
Annotations: Access project and catalog data to use as annotations in the VarSeq projects
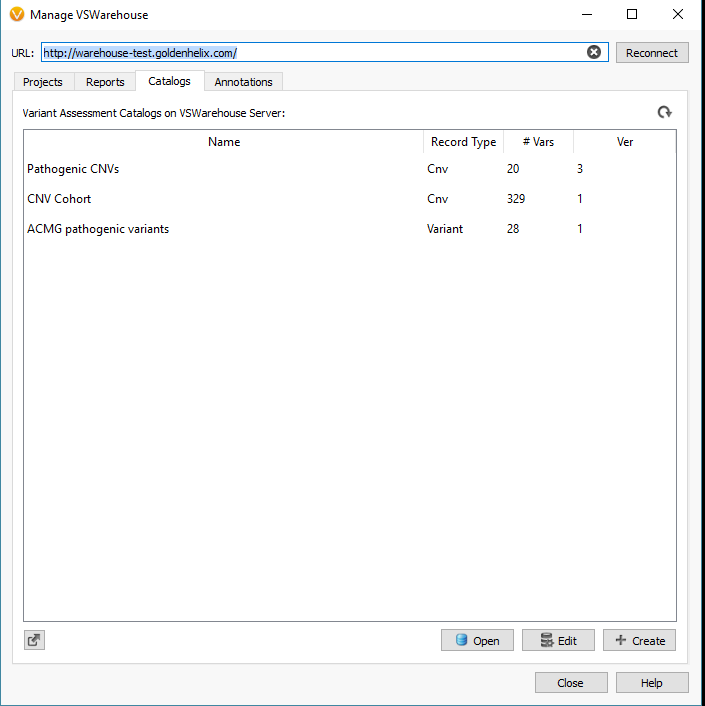
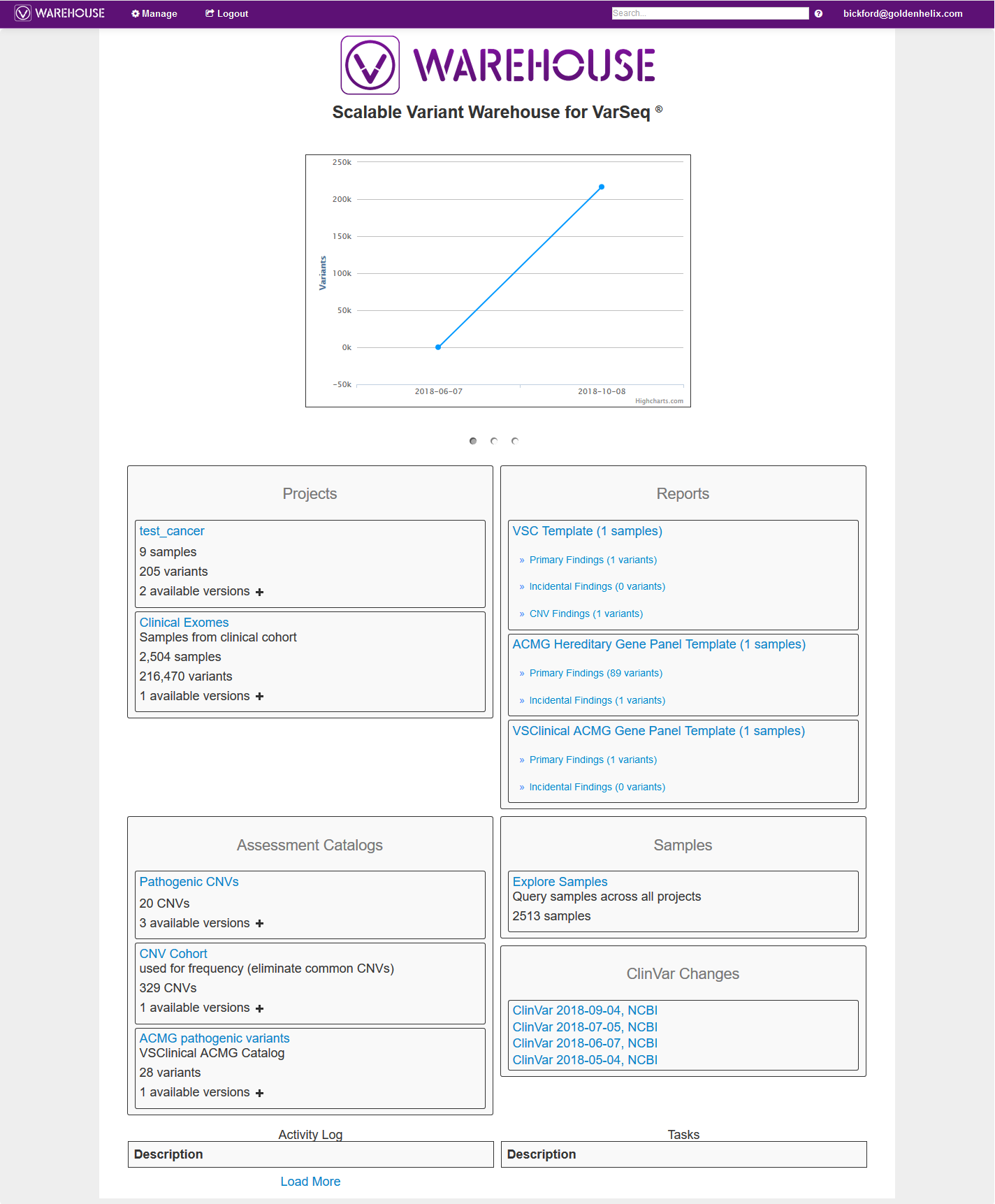
You will also notice the url at the top of Warehouse terminal; which will take you to the Warehouse browser seen in Figure 3. From this browser, the user can access projects, reports, assessment catalogs, manage permissions/user accessibility and track variant classification changes in ClinVar.
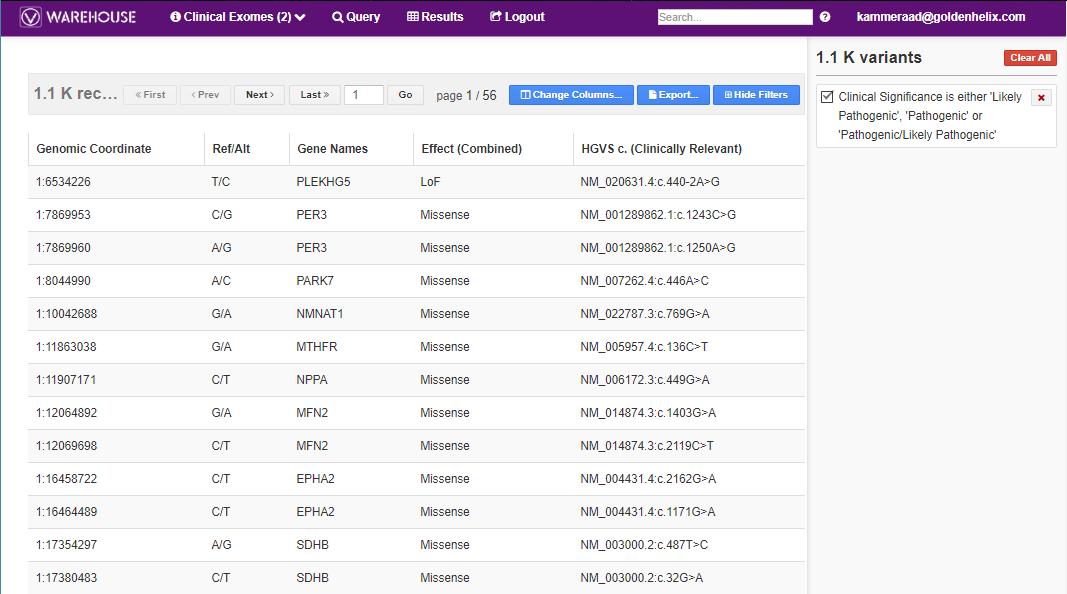
Users can do basic querying on their data across all the stored data in Warehouse. Using a Clinical Exomes project as an example in Figure 4, you can search through the VCF variant info, annotations, and algorithms in the project to filter on variants. After applying the filter on Query (seen on top of screen) the user then can access the Results to sift through the pathogenic variants present in the cohort project (Fig 5).
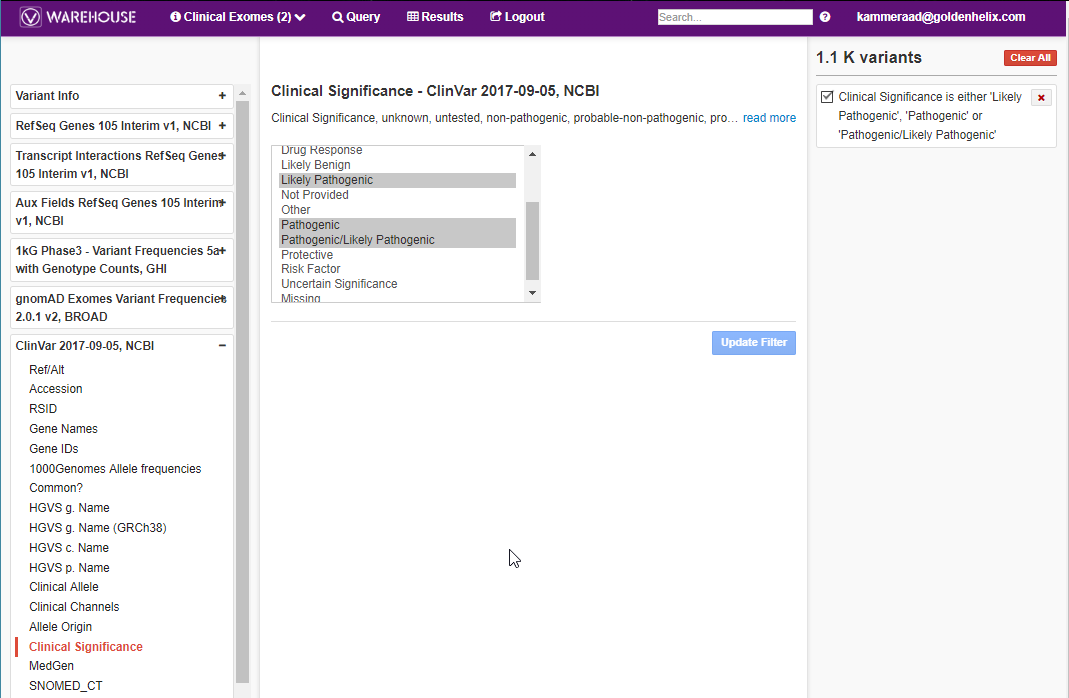
This filtering approach can also be utilized for assessment catalogs as well. In Figure 6 I am browsing through the VSClinical ACMG assessment catalog which was referenced in Part II of this blog series. In this search, I am looking for any variants in my catalog that are classified pathogenic/likely pathogenic.
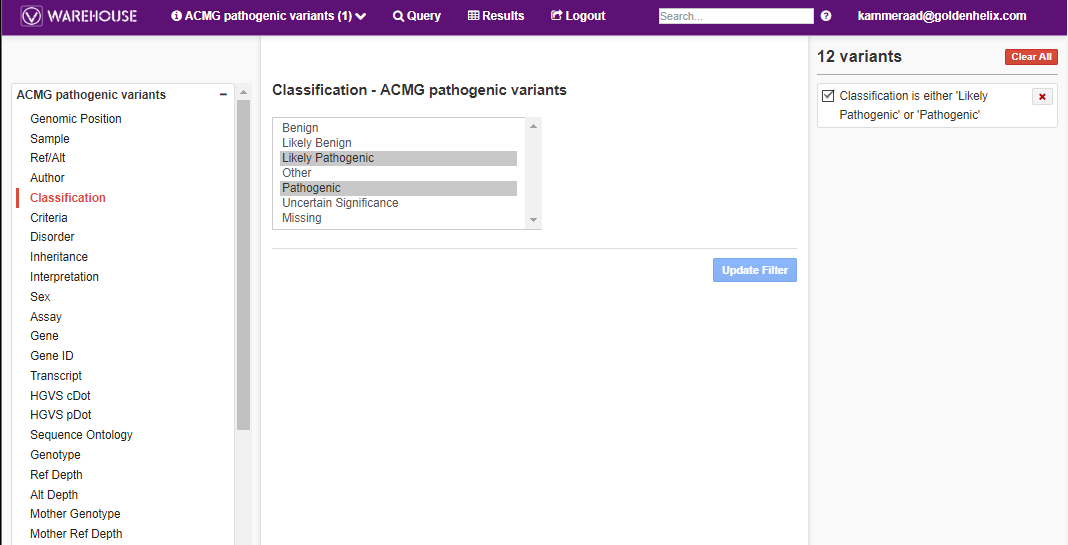
The results show not only each of the variants, authors who made their record and their interpretations (Fig 7).
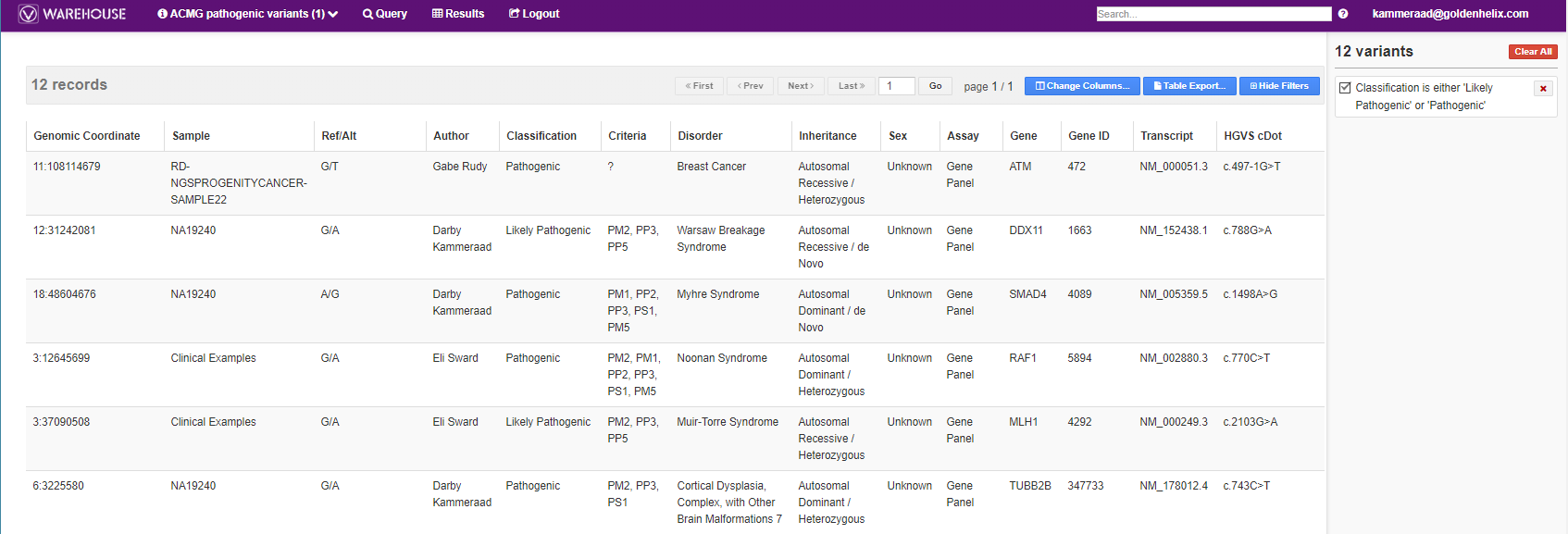
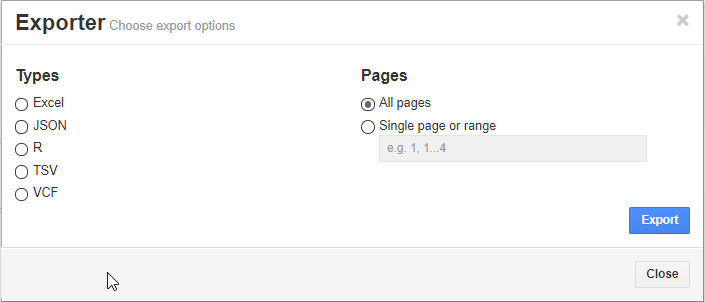
The user can also export the results in several different formats. One option may be to export the filtered variants into a VCF so to reimport into VarSeq.
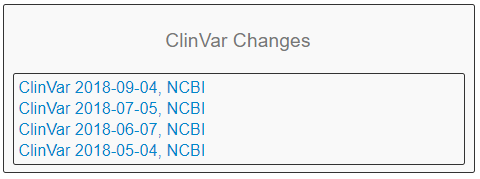
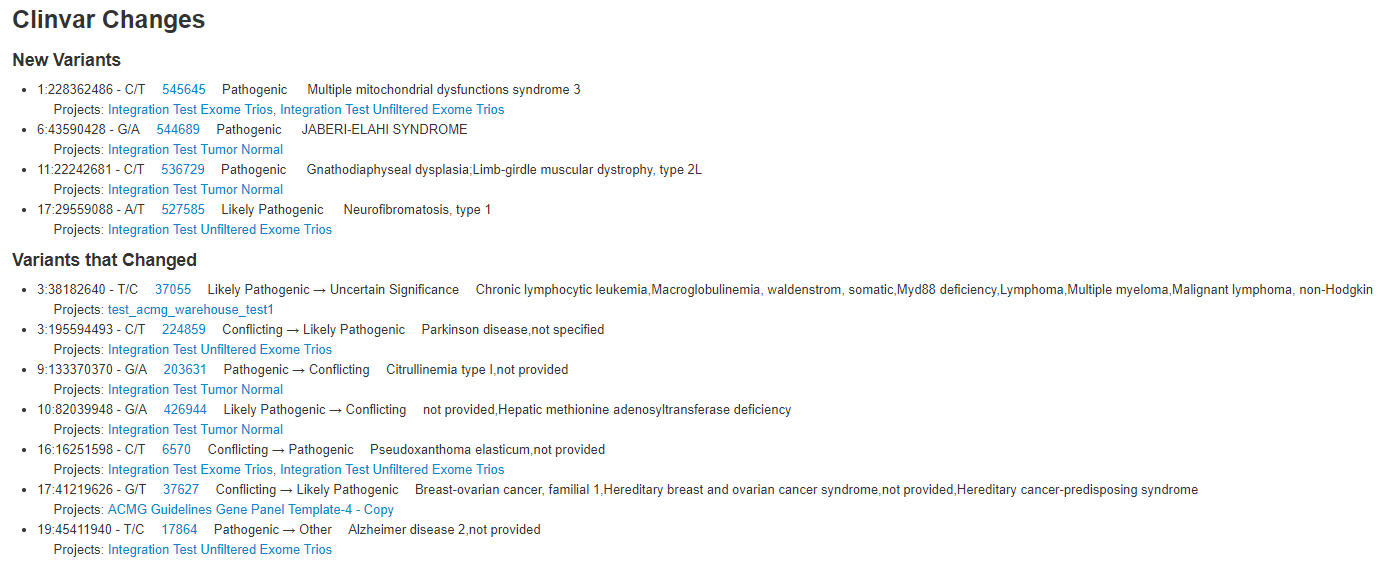
Another incredibly powerful tool in Warehouse is to quickly reference ClinVar classification changes. With the ClinVar Changes section seen in Figure 9, the user can see which variants are new to the most recent version of ClinVar. The user can quickly identify which variants that exist in their cohort are new to ClinVar or have changing classifications, but also quickly access the project/report/catalog that each variant exists in (Fig 10). With this tool, a user can see if some variants previously classified as benign or uncertain significance are now pathogenic, and directly browse to the project via hyperlink.
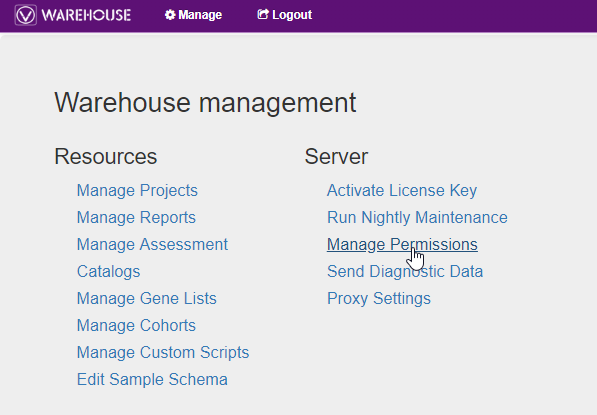
From the Warehouse browser, user accessibility can also be assigned. Settings and permissions can be accessed by clicking Manage in the top left corner of the browser (Fig 11). Not only can users manage their projects, reports, catalogs, but also modify the user permissions. This is important if you want to regulate some users to have full access to their projects, but only view data from other projects. An admin user would have full control to assign these permission customizations themselves.
Overall, the purpose of this blog series was to show the value of using new CNV and VSClinical stored data from VSWarehouse to optimize the analysis in VarSeq on new variants. The final part of this blog series focused on how to access the stored variants from the Warehouse browser/server. This was meant to be a brief overview of the capabilities to help orient users to the Warehouse/VarSeq interface, but also demonstrate the value of having cohort data in a central repository. With this exposure, we hope to peak your interest and spark the questions about what benefits Warehouse can provide to your analysis. We would love to hear from you and answer any of your questions directly through a one on one demo. Please feel free to email [email protected] with all of your questions.