In our latest VarSeq release, we updated our PhoRank algorithm with the ability to specify OMIM phenotype terms not present in HPO, as well as a general update to the algorithm to improve the results.
In this post, we review the fundamentals of how PhoRank determines the ranking of genes in your VarSeq projects based on your input phenotype terms (specified by project or by sample) and how our recent updates provide improved results over the original algorithmic strategy.
Revisiting PhoRank, Handling some Weaknesses in Phevor
In the spring of 2015, we released a phenotype driven variant ontological ranking tool called PhoRank. This tool was modeled on the Phevor algorithm developed by Mark Yandell’s group and works by leveraging the knowledge stored in biomedical ontologies, such as the Human Phenotype Ontology (HPO) and the Gene Ontology (GO). These ontologies model relationships between genes and diseases as directed acyclic graphs, where concepts are represented as nodes in the graph and the logical relationships between them are represented as edges.
The Phevor algorithm begins by first assembling a set of seed nodes. For each search term, its seed nodes are all nodes connected to it, either directly or through a shared gene relationship. Each seed node is then assigned a score of one, and these scores are propagated across the ontology as follows. First, the algorithm moves from each seed node toward its children, and each time an edge is traversed, the current value of the previous node is divided by two. This process continues until a leaf node is reached. Next, the seed scores are propagated upward toward the root nodes of the ontology in the same fashion. Finally, each node’s score is normalized, and each gene is assigned the score of its maximal scoring child node.
Recently we have constructed a new ontology based on terms stored in OMIM, which we have incorporated into the PhoRank algorithm. This means that OMIM terms can now be included in PhoRank queries and relationships between HPO and OMIM can be considered by the algorithm.
Let’s examine the results from the following query, which contains both OMIM and HPO search terms:
- OM:300257: Danon disease
- HP:0001685: myocardial fibrosis
- HP:0001324: muscle weakness
- HP:0100543: cognitive impairment
This query contains the OMIM term Danon disease along with several associated phenotypes in HPO. The results of this query are show in Figure 1.
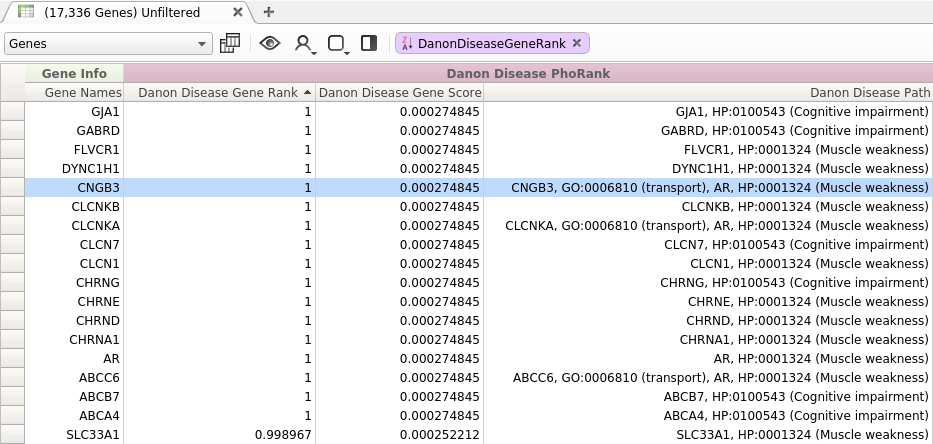
Figure 1: Danon disease Query
While most of the top ranked genes returned by this query are directly related to the entered search terms, there are a few top ranked genes that do not seem to be immediately relevant to the specified phenotypes. For example, one top ranked gene was CNGB3. This gene, which has no direct association with any of the entered phenotypes, scored higher than many genes which are directly related to the search terms. Why was this gene ranked so highly?
We can begin to answer this question by looking at the path reported by PhoRank:
- CNGB3
- GO:0006810 (transport)
- AR
- HP:0001324 (Muscle weakness)
This path passes through the term GO:0006810, which is a general term used to describe the movement of substance into, out of or within a cell. Because this term is so general, it is related to a large number of terms in the GO ontology. Specifically, this term has 351 neighbors! We will refer to such highly connect nodes as super nodes.
The Phevor algorithm tends to inflate the scores of super nodes, such as GO:0006810. Recall that the Phevor algorithm propagates scores from each node to its neighbors. Since super nodes have a large number of neighbors, they will receive a large number of scores during the propagation process. Even if the individual scores are very low, the sum of all propagated score values can be quite high. For example, even if the neighbors of GO:0006810 had scores as low as 0.01, the sum of the propagated scores would be 1.75. This means that GO:0006810 would have a higher score than even the initial seed nodes, despite having no direct relationship to any of the search terms!
A More Balanced Approach
To address this issue, we have modified the way our algorithm PhoRank sets the initial score of the seed nodes, so that very general unrelated terms, such as GO:0006810, will have a low initial score, while more specific nodes that are highly related to the search terms will have a high initial score. In the updated algorithm, each seed node is assigned a score of one, only if it is one of the initial search terms. All other seed nodes are assigned a score equal to their average similarity to the search terms. We measure the similarity between two nodes and as using the Jaccard index, which is defined as follows:
where of the number of nodes connected to the node [1]. The Jaccard index measures the similarity between two nodes and serves to decrease the score of nodes that are very general or have low similarity to their immediate neighbors. Similarity scores, such as the Jaccard index, have been used in the other phenotype-based prioritization algorithms to measure the relevance of ontology terms to user-specified phenotypes [1, 2, 3]. We have also modified the algorithm’s score propagation mechanism, so that the score propagated from one node to another is multiplied by the Jaccard index of the two nodes, thereby down-weighting the more general nodes in the path.
These improvements increase the scores of more specific nodes that are highly related to the search terms, while decreasing the scores of more general nodes with many neighbors. The effect of these changes can be seed when looking at the results in Figure 2.
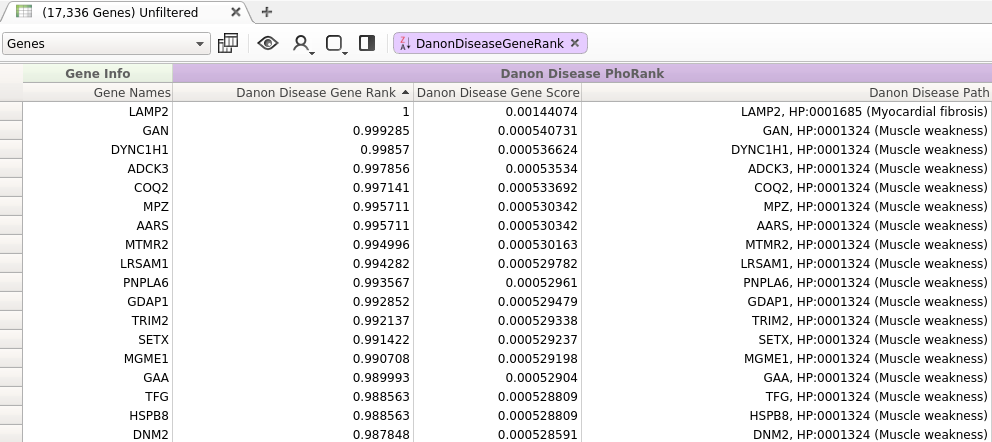
Figure 2: Query with New Algorithm
In contrast to the results presented in Figure 1, the top ranked genes returned by the modified algorithm all have a direct relationship to at least one of the entered search terms.
These improvements, combined with the ability to enhance your queries with OMIM, mean that PhoRank is now better than ever. If you want more information about PhoRank, or are interested in adding OMIM to your existing VarSeq license, please reach out to [email protected]. Our team of experts would be happy to demonstrate how PhoRank can be incorporated into your existing workflows.
References:
[1] Smedley, D., Oellrich, A., Köhler, S., Ruef, B., Westerfield, M., Robinson, P., … & Sanger Mouse Genetics Project. (2013). PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database, 2013, bat025.
[2] Köhler, S., Schulz, M. H., Krawitz, P., Bauer, S., Dölken, S., Ott, C. E., … & Robinson, P. N. (2009). Clinical diagnostics in human genetics with semantic similarity searches in ontologies. The American Journal of Human Genetics, 85(4), 457-464.
[3] Masino, A. J., Dechene, E. T., Dulik, M. C., Wilkens, A., Spinner, N. B., Krantz, I. D., … & White, P. S. (2014). Clinical phenotype-based gene prioritization: an initial study using semantic similarity and the human phenotype ontology. BMC bioinformatics, 15(1), 248.