It may have been easy to miss in the drum-beat of monthly annotation updates we do here at Golden Helix, but there are a couple of things that are very special about the January update to the ClinVar database:
- We added new fields including HGVS names of variants and citations in PubMed for variants
- ClinVar nearly doubled in size by a massive submission of clinical assertions from Illumina Clinical Lab!
From Hobbyists to Your Clinic: Illumina’s UYG Benefits All
Back in 2009, the advent of Whole Genome Sequencing as an economically viable test to end all tests was reaching a fever in the medical genetics world.
But our understanding of the genome outside a select well-studied set of genes was limited. The task of interpreting the multitude of Variants of Unknown Significance scattered across an exome, let alone a whole genome, seemed nearly insurmountable. Most clinical labs decided to start off their genetic testing offerings with gene panels, considering WGS too much to take on.
Illumina decided to take things into their own hands with the establishment of their own fully funded and staffed CLIA certified Illumina Clinical Services Laboratory.
The lab had one test on the menu: Whole Genome Sequencing
Being a trailblazer requires laying down your own tracks. To help understand the baseline of clinical grade human variant annotation, they bootstrapped their lab processes and internal databases by being the service behind the yearly Understand Your Genome conference.
In this cleverly designed win-win, early adopters, curious medical professionals and many industry patrons pay 5K to attend the Understand Your Genome conference held annually in San Diego (Illumina’s back yard). Conference goers show up, are handed their fully sequenced and analyzed whole-genomes at the door and listen to talks given by experts and medical professionals discussing the state of the art in genome interpretation.
By 2015, Illumina had sequenced over 627 whole genomes, each carefully analyzed using the ACMG guidelines with each unknown variant being reviewed using bioinformatics and manual literature curation along with close interpretation of the (often healthy) patient’s medical history, doing genetic counseling as necessary to follow up with results.
Maybe surprisingly, 38% of healthy individuals had results that are expected to be clinically significant. Although the majority of these findings are in common, low-penetrant conditions, some were in actionable highly penetrant diseases.
As you can imagine, the Variant Scientists at the Illumina lab have made many variant classifications over the year, discovering many novel Pathogenic or Likely Pathogenic variants as they process these whole genomes of both healthy and diseased individuals.
They have also marked many existing, literature curated “Pathogenic” variants to “Variants of Unknown Significance” or “Likely Benign” as they observe these variants in many healthy middle age adults with no sign of the penetrant diseases they are proposedly associated with.
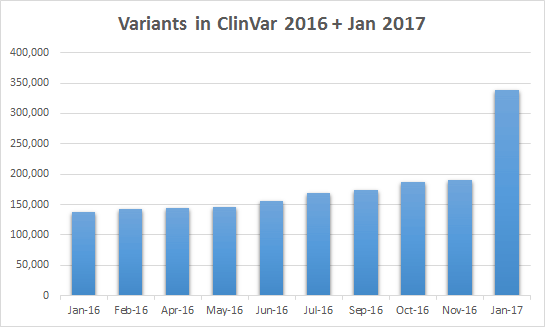
In the January release of ClinVar, Illumina has taken the step to submit back to ClinVar what I assume is the full contents of its clinical variant assertion database!
ClinVar is built by the collaborative submissions of many clinical labs, both commercial and academic.
With this one submission, Illumina is now the single largest submitter (by four times) to the ClinVar database!
Well done Illumina!
More Variants, and Also New Fields
Along with this jump in variant assertions, we also expanded our curation of ClinVar to pull in a couple other useful fields for each record.
| HGVS g. Name | Variant names from HGVS. The order of these variants corresponds to the order of the info in the other clinical INFO tags. |
| HGVS g. Name (GRCh38) | Variant names from HGVS for the opposite build |
| HGVS c. Name | HGVS c. nucleotide expression |
| HGVS p. Name | HGVS p. protein expression |
| Citations | Citations from PubMed and NCBI articles. |
These ClinVar provided HGVS names may be useful for pulling directly into a report or grabbing the position of the variant on the GRCh38 genomic coordinate system for reference.
The Citations field provides hyperlinked article IDs to PubMed and other NCBI written resources about diseases that provide evidence for the current variant assertion.

As always, Golden Helix is committed to providing the best possible curation of the many complex and scattered public data sources in one streamlined and updated repository, available for use in your variant annotation and interpretation workflows in VarSeq or your research workflows in SVS.

how do I access this new information?
You probably figured it out by now. Inside VarSeq or GenomeBrowse you can select >Tools >Data Sources .. Click on Public Annotations and scroll down to ClinVar 2017-01-6, NCBI Select the track and click to Download.
Comment: Great.!!!
Now please me as to how to access this new information.