Following up on our recent post about VSWarehouse 3’s Bring Your Own Cloud capabilities, we wanted to dive deeper into one of its most powerful features: our comprehensive workflow system. This system is designed to streamline genomic analysis pipelines while providing flexible integration with various cloud genomics providers.
Understanding VSWarehouse 3 Workflows
At its core, VSWarehouse 3’s workflow system is built around the concept of modular, configurable tasks that can be combined to create end-to-end analysis pipelines. Each workflow is composed of individual tasks that handle specific aspects of data processing, analysis, and reporting. This modular approach allows labs to create standardized, reproducible processes while maintaining the flexibility to adapt to different analysis requirements.
Task Integration with Cloud Providers
A key strength of our workflow system is its ability to integrate with any cloud genomics vendor that provides access through API. Through our task-based architecture, VSWarehouse 3 can:
- Pull Data from Cloud Providers:
- Automated data retrieval from platforms like BaseSpace and Archer Dx
- Direct integration with vendor APIs for seamless data access
- Support for various data types and formats, including VCFs, CRAMs, and BAMs
- Process Analytics Within Workflows:
- Standardized analysis pipelines for secondary analysis using Sentieon base pipelines
- Support for custom genomics pipelines built using any software that can run on Linux
- Automation of VarSeq annotation and reporting workflows with VSPipeline
- Custom parameterization based on sample and data configuration
- Push Results to External Systems:
- Integration with laboratory information management systems (LIMS)
- Support for the creation of downstream population-based annotation sources
- Data archiving and transfer to long-term storage
Real-World Applications
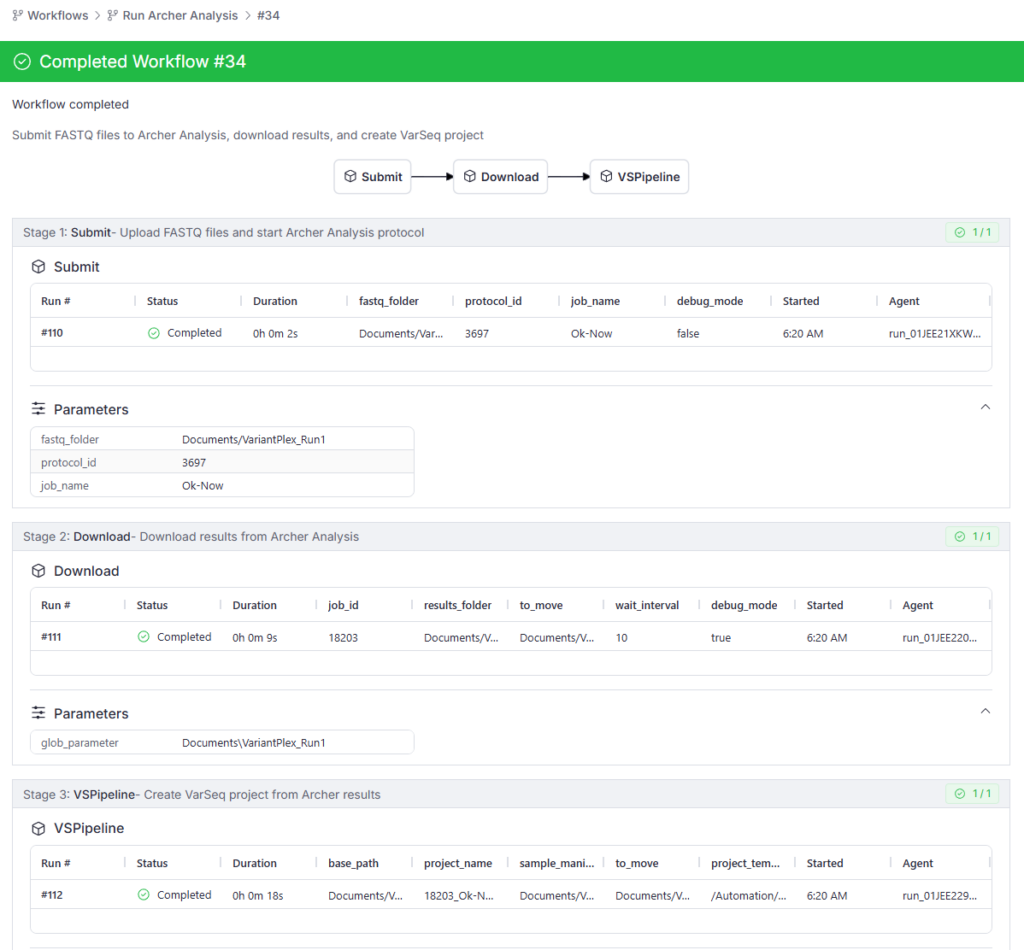
For example, a typical workflow might include:
- A task that uploads to Archer the FASTQ files from a sequencing run and starts an analysis protocol
- A download task that waits for the Archer Analysis job to complete and downloads the results
- A project creation task using VSPipeline to annotate, filter, and prepare a preliminary report
Looking Forward
We continue to expand our integration capabilities based on user needs. Whether you’re working with BaseSpace, ArcherDx, or other cloud genomics providers, VSWarehouse 3’s workflow system can help streamline your analysis pipeline. It will allow seamless automation, integrating these third-party systems with your existing VSPipeline and VarSeq workflows.