Compound heterozygosity describes the relationship between two alternate alleles when they are located within the same gene but at different loci within that gene. Compound heterozygosity is particularly relevant in a recessive disorder when the presence of these alleles in combination confers an increased risk of disease, similar to a traditional homozygous recessive combination of alleles. The detection of compound heterozygosity between single nucleotide polymorphisms (SNPs) and copy number variants (CNVs) when analyzing NGS data can be a laborious task without the proper bioinformatic tools to automate this process. Yet, the ability to detect these events is crucial to the understanding of the interactions between variant types and promises to improve the diagnostic and therapeutic yield of genetic analyses. So, we are pleased to announce that in VarSeq 2.6.2, users will have the added capability to analyze compound heterozygosity between single nucleotide variants and copy number variants.
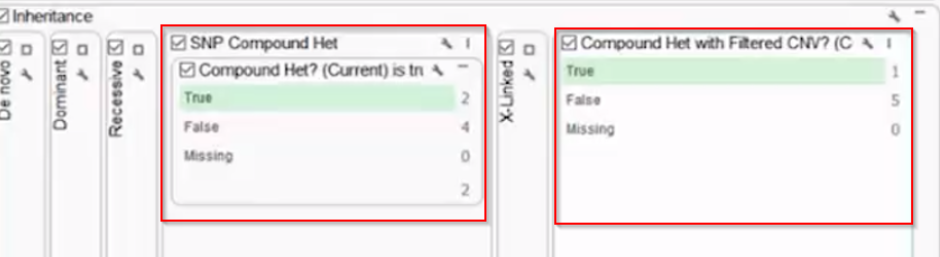
Previously, VarSeq’s compound heterozygosity algorithm only detected compound heterozygotes between small variants (SNPs and indels). Our new algorithm will expand this detection capacity to copy number variants as well. This includes CNVs detected in VarSeq or imported from another secondary caller (Figure 1). Users can now detect small variants in compound heterozygosity with CNVs or CNVs with each other.
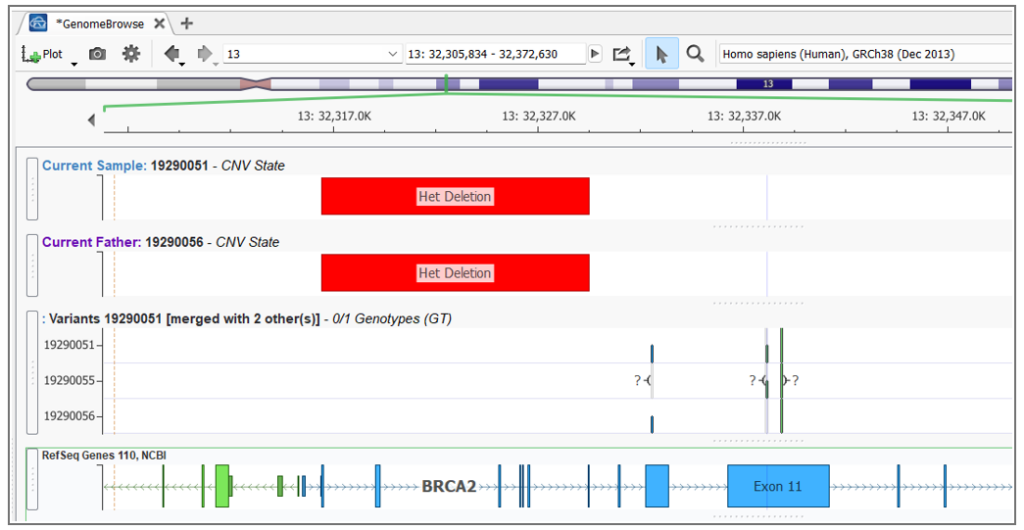
The example below is from a family analysis in which the proband has inherited a BRCA2 SNP from their mother and a BRCA2 CNV from their father (Figure 2). This is a clear instance of compound heterozygosity, which can be detected by visual inspection of calls in the project and perusing the location of the different variant types in relation to each other in Genome Browse. However, that is a very cumbersome approach. Now, with the *NEW* Compound Heterozygosity detection algorithm, detecting such events is a breeze!
To see this new algorithm in action, check out our webcast: Combined Impact: New Tools to Assess Complex and Compound Heterozygous Variants with VarSeq! This webcast highlights our new tools to assess compound heterozygosity, as well as several other complex genomic events.