Thank you to all our audience members who attended our recent webcast, Combined Impact: New Tools to Assess Complex and Compound Heterozygous Variants with VarSeq. If you would like to view the webcast, follow the link above! As the title suggests, this webcast was all about breaking down the new variant analysis tools in the upcoming VarSeq 2.6.2 release and highlighting our current tools for methylation analysis.
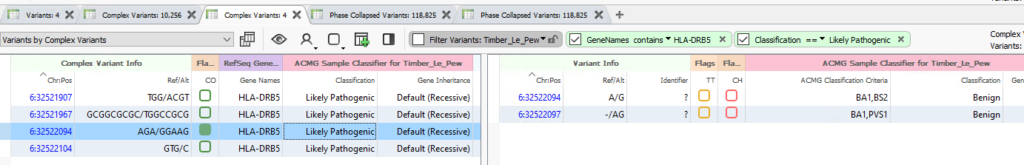
Starting with our new tools, one of our major upgrades includes changes to our data importing. Previously, if a user wanted to view variants both in their native complex state or as a variant split into allelic primitives, the user would have to do multiple projects with different import options. Those days are behind us with the new Complex Variant Tables. As seen in the webcast, we are now able to see the complex variant side-by-side with the allelic primitives state (Figure 1). This gives the user full control over which variants to import into VSClinical for further analysis.
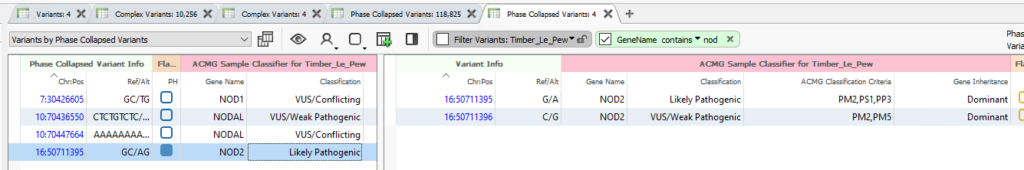
Our next major upgrade is the Phased Collapsed Variants Table. By leveraging the phased status of nearby variants, we can collapse multiple variants into one variant record for a more accurate assessment. Here, we have an example in NOD2 where individually, the records receive a VUS and a Likely Pathogenic score, but together, they get a Likely Pathogenic score (Figure 2).
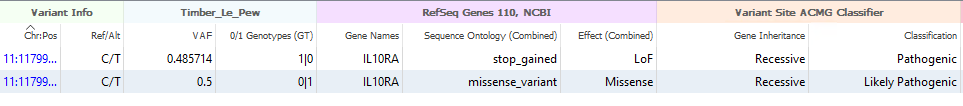
In addition to the Phased Collapsed Variants algorithm, we have another new algorithm that also leverages the phased condition of a set of variants, but in a different way. This algorithm is the Phased Compound Heterozygous Variant algorithm, which, as the name implies, assesses the GT position of variants in both chromosomes to report Compound Het variants. Here, we have an example of two variants on opposite strands of chromosome 11 (Figure 3). Both are in the Recessive gene IL10RA, with Pathogenic and Likely Pathogenic scores. These Compound Het variants would then be taken into VSClinical for further analysis. This is a unique algorithm in that, previously, you would need a Trio setup (Proband, Mother, and Father) to classify a variant as Compound Het. Through the use of long-read sequencing and phasing, we are able to make this assessment in a single sample project.
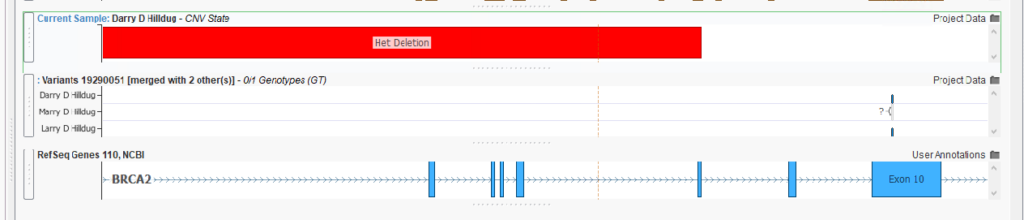
The last new algorithm we showcased in this webcast has been a customer request for quite a while. We are thrilled to announce, with the new 2.6.2 release, the ability to identify SNP and CNV Compound Het variants in a trio! This algorithm, appropriately called the CNV Compound Het Detection Algorithm, will leverage inheritance information from both of the parents to identify genes that have been affected on both chromosomes by both a small variant (SNPs, INDELs, etc.) and a larger copy number variant. Here we see an example in BRCA2, which has both a Het Deletion and a pathogenic small variant inherited from a parent (Figure 4).
All of these new algorithms introduce a new level of analysis control for our users. We are excited to see what new workflows our users develop now that they will have more fine-tuned control over viewing these complex variant states.
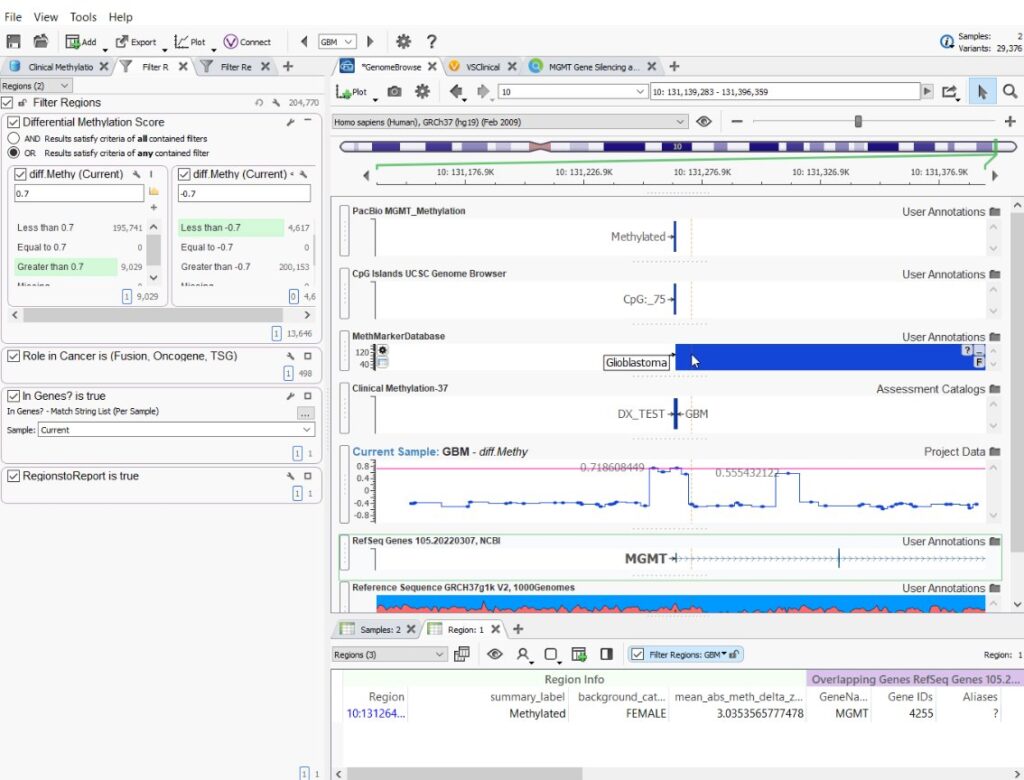
In addition to our new tools, our webcast also covered how VarSeq can be used to assist in the evolving field of Methylation Analysis! We are excited about the new developments in the field of methylation research. As more of our customers turn to long-read sequencing and have access to differentially methylated region data, we are developing new capabilities and tools to assist in this area of analysis. We already have the ability to plot, filter, and create custom biomarker interpretations for final reporting (Figure 5). As we look to expand our tools in this area, feel free to reach out to [email protected] and we can chat about how methylation data is used in your diagnostic workflow.
We had several questions we were not able to answer in our webcast that we are happy to talk about here!
Question 1: We saw a webcast about the PGx add-on feature a few months ago. Does this version of VarSeq have any new tools in this area?
Answer 1: Yes! We have another webcast coming in October 2024 that will showcase the new features and databases for our PGx workflow.
Question 2: Are you going to have any methylation-specific databases?
Answer 2: We are adding a database called MethMarkerDB. If you would like to see any specific databases, please let us know at [email protected], and we can chat about them.
Question 3: Can the complex variants be added to my current assessment catalogs, or do I need to make all new assessment catalogs?
Answer 3: You will be able to add the complex variants to your current assessment catalogs and evaluate them in VSClinical like you would the allelic primitive variants.
Question 4: Does the CNV and small variant compound het algorithm work on a single sample or just a trio?
Answer 4: To clarify here, we have added two new compound het algorithms. The first is small variant-specific, and it can leverage phasing information to determine if two small variants are in compound het for the same gene. This algorithm does the work that previously could only be done in a trio. The other algorithm looks at compound het for small variants and CNVs. This is an all-new capability, but it requires the mother and father samples to look for the inheritance of the CNV and the small variant.
Thank you for the great questions, and we will see you on the next webcast!