Our customers asked for a step-by-step tutorial for navigating a pharmacogenomic (PGx) workflow in VarSeq, and we are happy to have it delivered! Our brand new PGx tutorial can be found on our website, along with our other VarSeq tutorials. You can also follow this link. Like our other famous tutorials, this one will provide example data, or you may follow along with your own star allele called data. This PGx tutorial breaks down the importance of star allele calling, how to use CPIC to determine PGx diplotypes, and how to use a text manifest to report on a patient’s medication recommendations. Moving all the way through the PGx analysis process, this tutorial concludes with rendering a report for our patient, John Doe (Figure 1).
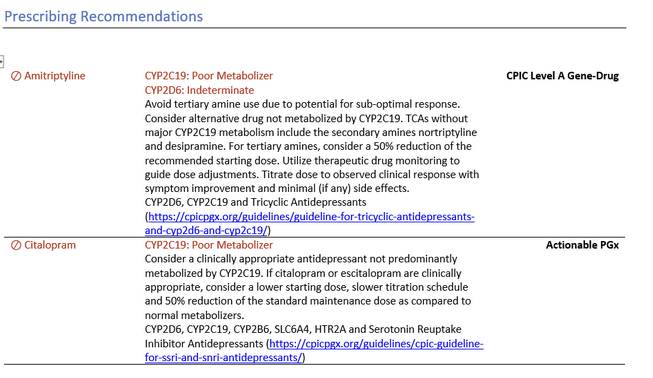
As to be expected with a Golden Helix report, our PGx report contains a wealth of information for our patient, including personalized prescribing drug recommendations based on his gene diplotype (Figure 2). By leveraging his diplotypes to define his metabolizing phenotypes, we can look for actionable recommendations both for a current medication list, and medications our patient may encounter in the future. The PGx Module will also be able to break down a comprehensive list of Alleles Tested and Alleles Not Tested, for assurance that we have accounted for all the possible diplotype combinations for metabolizer phenotypes.
To go through all of these steps and more, check out our new PGx Tutorial today! For more information about our new VarSeq PGx Module, you may want to check out our VarSeq 2.6.0 Pharmacogenomics webcast or our PGx User Perspective webcast. As always, if you have any questions about how our PGx Module can be used to complement your unique use case, please send an email to [email protected] or visit our VSPGx product page to learn more and request a demo.