While the analysis of gene fusions is crucial for understanding the genetic basis of cancer, the process of interpreting these mutations can be challenging. One important component of fusion interpretation is the identification of relevant publications. To aid researchers in the search for publications related to specific gene fusions, Felix Mitelman and colleagues have created the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer [1].
This database is the result of a pioneering effort to catalog data related to chromosome aberrations and gene fusions observed in various cancer types. Driven by the motivation to integrate fragmented knowledge in the field, Mitelman recognized the importance of consolidating data from various sources and curating it into a freely accessible repository for the clinical and scientific community.
Mitelman and his colleagues have painstakingly poured over the literature and cataloged publications discussing specific gene fusions as they relate to various forms of cancer. The database currently boasts an extensive repository featuring over 33,000 unique gene fusions spanning more than 14,000 genes. In 2019, Denomy et al.demonstrated the extensive nature of the Mitelman database in a systematic comparison against the Cancer Genome Atlas (TCGA), where the authors found that the Mitelman database provided unique information not captured by the population-based genomic analyses used in TCGA [2].
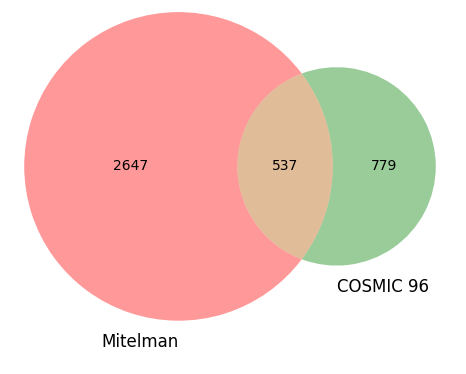
We performed our own comparison of the Mitelman database against the COSMIC 96 database of gene fusions [3]. The Venn diagram below shows overlap between the PubMed references from each database.
The Mitelman database boasts an impressive 3,184 PubMed citations directly related to gene fusions in cancer, of which only 537 were found to be shared with COSMIC 96. This indicates a significant disparity in the number of gene fusion citations between the two databases. In fact, COSMIC 96 contains fewer than half the total number of gene fusion citations present in the Mitelman database, with only 779 citations being unique to COSMIC 96.
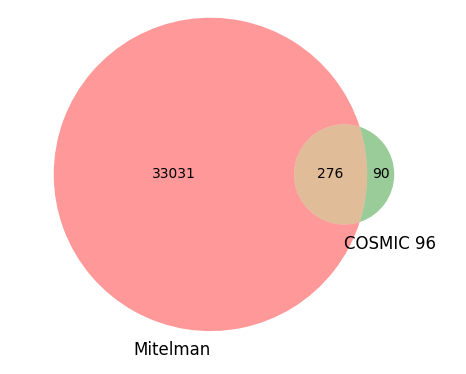
The disparity between the two data sources becomes even more striking when you compare the number of unique fusions in each source.
As stated above, the Mitelman database contains over 33,000 unique gene fusions, of which only 276 are shared with COSMIC 96. Conversely, COSMIC 96 contains a mere 90 unique fusions that remain exclusive to its database and are not found in the Mitelman repository. This comparison underscores the impressive breadth of the Mitelman database, demonstrating its significance as an indispensable resource for identifying publications linked to specific gene fusions.
To help our users leverage this powerful resource, Golden Helix released the Mitelman Fusion Pairs annotation track for VarSeq. Using this track, fusions imported into VarSeq can now be annotated against the Mitelman database, allowing users to quickly identify publications associated with imported fusions. If you have any questions about this new annotation track or about fusion support in VarSeq, we encourage you to reach out to us at [email protected].
References
- Mitelman, Felix. “Mitelman database of chromosome aberrations and gene fusions in cancer.” https://mitelmandatabase.isb-cgc.org/ (2010).
- Denomy, Connor, et al. “Banding together: a systematic comparison of the cancer genome atlas and the Mitelman databases.” Cancer Research 79.20 (2019): 5181-5190.
- Tate, John G., et al. “COSMIC: the catalogue of somatic mutations in cancer.” Nucleic acids research 47.D1 (2019): D941-D947.