In our recent webcast, we discussed the exciting new features of VarSeq 2.4.0 and the updated VSClinical interface. The discussion was centered around three main topics:
- Significance of Structural Variants: We emphasized the vital role of structural variants in analysis of cancer and diagnosing rare diseases. This included reviewing the different types of structural variants and how the current advances in long-reads and other technologies provide the opportunity to create a single comprehensive NGS test.
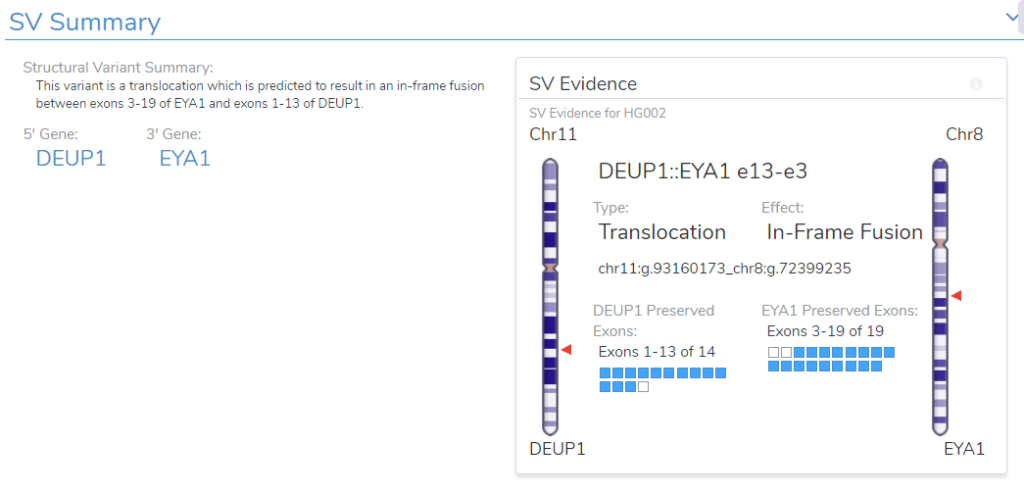
- Structural Variant Interpretation: We reviewed how the analysis and interpretation workflow can include CNVs and structural variants along with small variants. For structural variants, this begins with the genomic effect analysis done on the “break-end” table and ends in the interpretation and reporting done in VSClinical.
- Advanced Automation: A major highlight of the webinar was the advanced automation capabilities within VarSeq 2.4.0. We showcased the application of evaluation scripts and VSPipeline for automating steps in the analysis workflow. This enhancement reduces the risk of errors and improves efficiency, aiding faster and more accurate test results.
In summary, VarSeq 2.4.0 uniquely supports the analysis of all variant types in the clinical interpretation workflow. By incorporating structural variants, enhancing automation, and empowering users to handle complex data, it offers a comprehensive solution for diagnosing Mendelian disorders.
The webinar concluded with a Q&A session, where we answered several great questions:
- Can VarSeq 2.4.0 support fusions called from Ion Torrent data? Yes, it supports all input data platforms. There are specific built-in importers for fusion data from Ion Torrent and ArcherDx.
- How long does it take to upload samples to Golden Helix and get results? Our on-premise solution’s processing time is dependent on the available compute resources. Smaller panels can be processed in minutes, while larger panels and whole genomes may take around an hour per sample. It is suggested to use the capabilities of VSPipeline’s batch-oriented automation in this case so the annotation work is done before the interpretation starts.
- How is the pathogenicity of germline fusions determined? The pathogenicity is evaluated based on the resulting protein product of the fused exons. A deeper interpretation involves understanding the gene’s impact and role in disease, which is supported by numerous gene-level data sources presented in the VSClinical interface.
Stay tuned for more webcasts, where we will explore in greater detail the advancements in variant interpretation and automation offered by VarSeq and VSClinical.