Discover the latest advancements in cancer genomic profiling with the release of VarSeq 2.3.0
We are very excited to announce the release of VarSeq 2.3.0! This release was one of the largest VarSeq releases yet, as it includes a large refactor to the VSClinical AMP cancer module. A primary motivation for the release was focused on the availability and increased application of the cancer genomic profiling (CGP) kits made available by several precision oncology NGS solutions. Over the past few months, we have hosted a number of webcasts that have introduced many of the new features in VarSeq 2.3.0. However, I want to take a moment and break down some of the larger feature milestones that all users of the VarSeq and the cancer workflow should know about and take advantage of.
When you open VSClinical AMP, each tab across the top has been restructured to facilitate analyzing biomarkers from a clinical perspective. The Clinical Evidence tab merges the biomarkers and clinical trials tabs so available drug therapies, diagnostic or prognostic information, and associated clinical trials for variants that have been added to the evaluation are analyzed in one central location. The most noteworthy updates to VSClinical AMP are:
- A new curation interface is now available wherein you can write your own cancer interpretations that can be saved and reused in evaluations. Previously, to edit or write new interpretations, an evaluation needed to be created first for a sample. This feature is available as a third dropdown option, “Cancer Interpretations,” when opening VSClinical.
- The following annotation sources have been added to VSClinical AMP: NCT Trials, Drug Central, CIViC Assertion Summaries, and NCI Thesaurus Drugs. The following sources have been removed: PMKB and CGI Biomarkers. The following sources have been significantly updated: Golden Helix CancerKB and Cancer Ontology.
- There is a new evaluation option to allow setting interpretation match behavior to “Match Best,” Match All,” and “Match All Including Other Tumor Types.”
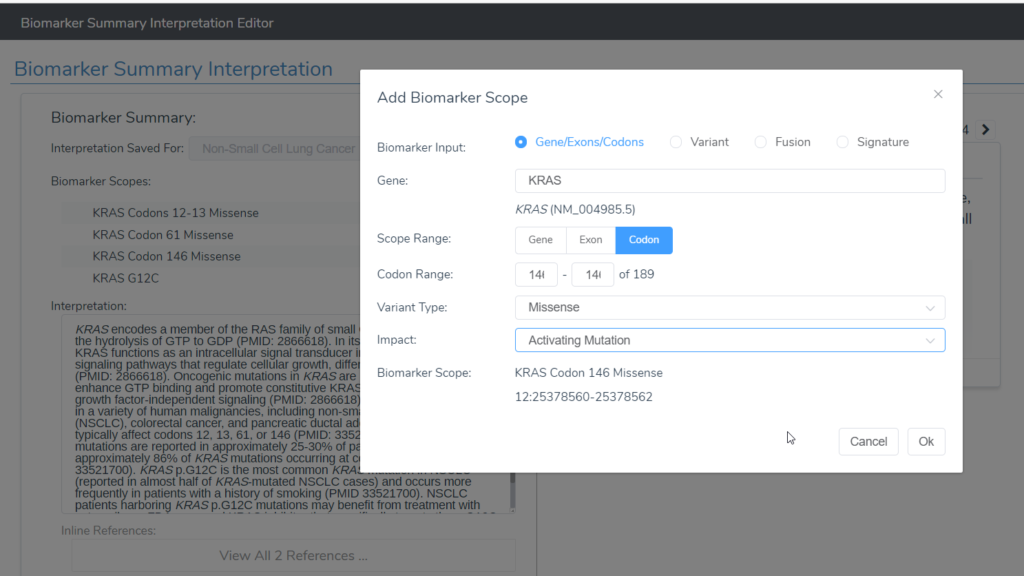
- VSClinical AMP now supports saving biomarkers as variants at a region (codon or exon range) or at a gene level. In addition, the variant impact can now be associated with biomarkers within VSClinical AMP: “Activating,” “Gain of Function,” “Loss of Function,” “Loss,” “Negative Finding,” and “Unknown” (Figure 1).
- The clinical trials search dialog allows users to search by drug, biomarker, and disease and these results can now be filtered by trial phase, patient age, patient sex, region (such as Europe) or country, and by distance using United States and European postal codes.
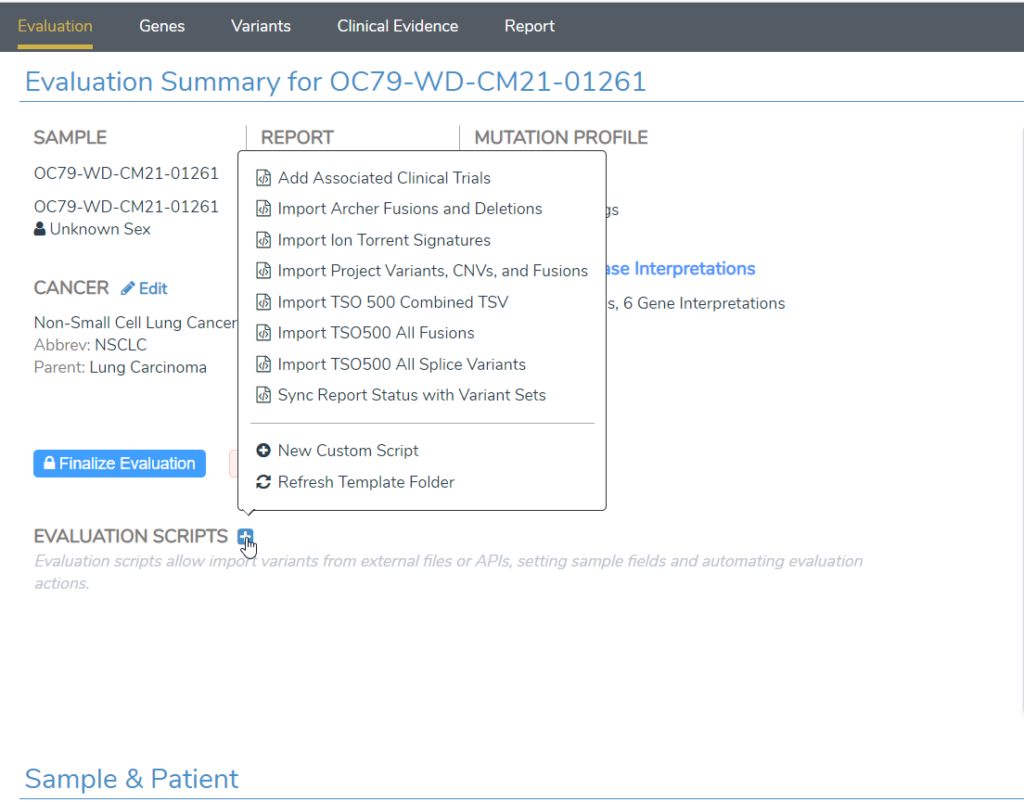
- VSClinical AMP now has “Evaluation Scripts” on the first screen that enable importing of custom data files as well as automation of many capabilities of the workflow. Evaluation scripts can also be called from VSPipeline to automate many tasks that previously required manual user action (Figure 2).
Outside of VSClinical AMP, there are some big features worth highlighting as well.
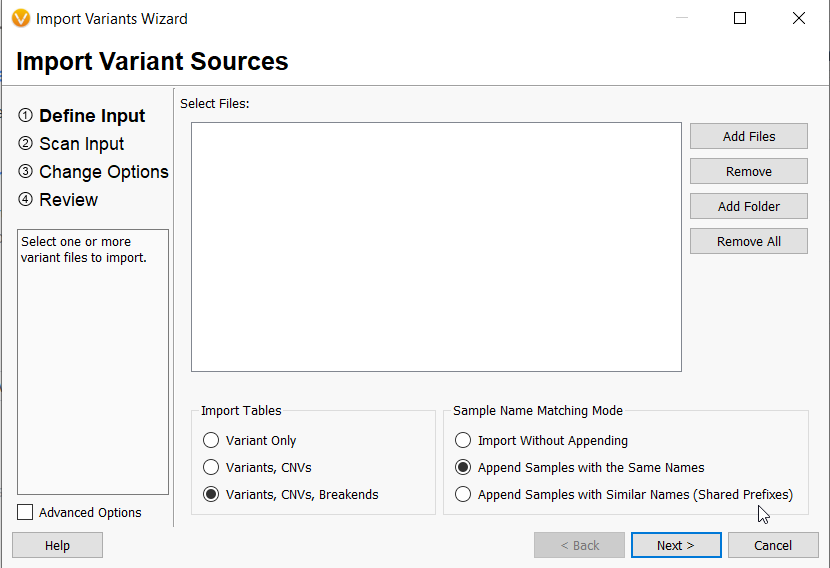
- VarSeq can now import either a single vcf file or multiple vcf files for one sample containing not only small variants but also copy number variations and structural variants (Figure 3). In addition, new algorithms and annotations have been made available for SVs, given this new capability.
- Variant import can be filtered based on fields from the SNV, CNV, and SV vcf files, such as filter = PASS, alt allele frequency, or read depth.
- CRAM files can now be imported into VarSeq, used for computation, and visualized within Genomebrowse.
- There is now support for the T2T CHM13 genome assembly.
- A new Comprehensive Cancer Template is now available for project creation, and 3 new report templates for VSClinical AMP.
Believe it or not, I have only mentioned a handful of the new features and updates within VarSeq 2.3.0. If you are interested in browsing the complete list of features and bug fixes for the 2.3.0 release, I encourage you to check out the VarSeq 2.3.0 Release Notes page. In addition, check out some of the recent blog posts and webcasts that discuss the changes and updates in detail. Please reach out to [email protected] if you have questions about any new features or need assistance upgrading to VarSeq 2.3.0!
