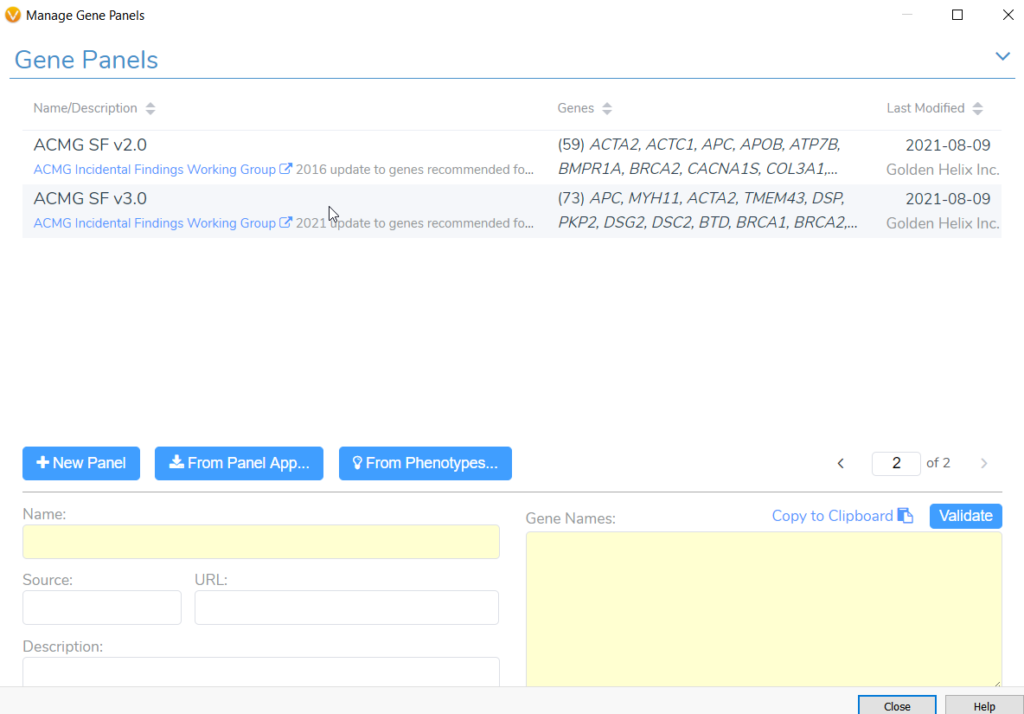
Tumor profiling via next generation sequencing (NGS) often reveals secondary germline variants that may constitute important incidental findings. In May 2021, the American College of Medical Genetics and Genomics (ACMG) released an updated policy statement for reporting incidental findings in exome and genome sequencing data along with a corresponding list of genes. These recommendations state that laboratories should report pathogenic and likely pathogenic mutations occurring in genes on the secondary findings list, because these genes are linked to “actionable disorders”. In our Gene Panel library (Figure 1), VarSeq houses both versions of this list, namely version 2.0 with 59 genes and the more recent version 3.0 with 73 genes. Any user doing clinical variant analysis in a cancer setting can take advantage of these gene panels to keep track of notable secondary germline variants that may be present in a cancer patient. A common challenge for users doing somatic variant analysis is how to include suspected germline variants as part of a somatic clinical report in keeping with the AMP guidelines. In fact, this question was posed during one of our recent webcasts on customizing clinical reports. So, in this blog we will briefly review how to report secondary germline variants in VSClinical AMP.
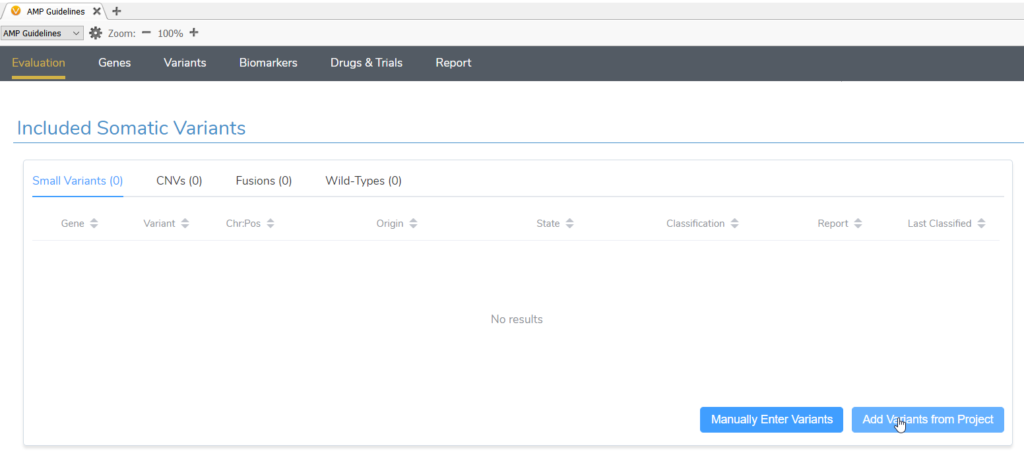
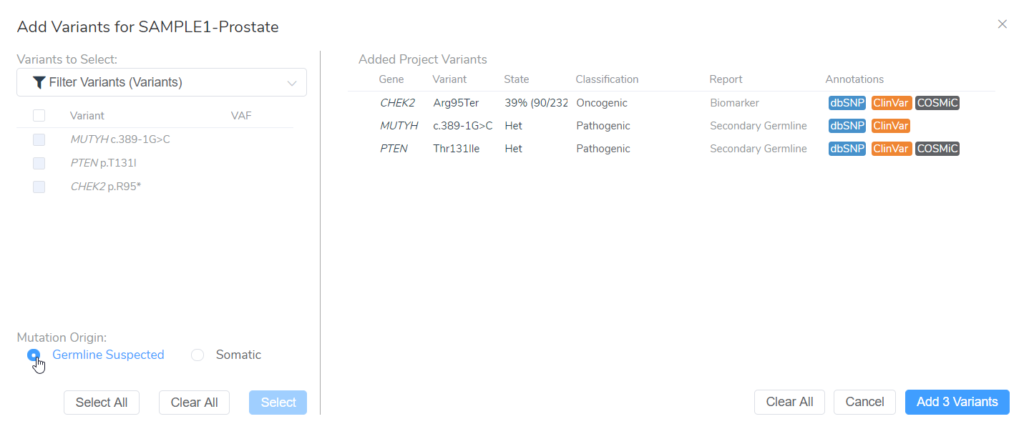
Once suspected secondary germline variants are identified, these can be brought in for analysis in VSClinical AMP alongside any somatic variants identified in the same project. After starting a new evaluation in VSClinical and selecting the tumor type, both somatic and germline suspected variants should be imported in the Included Somatic Variants tab (Figure 2).
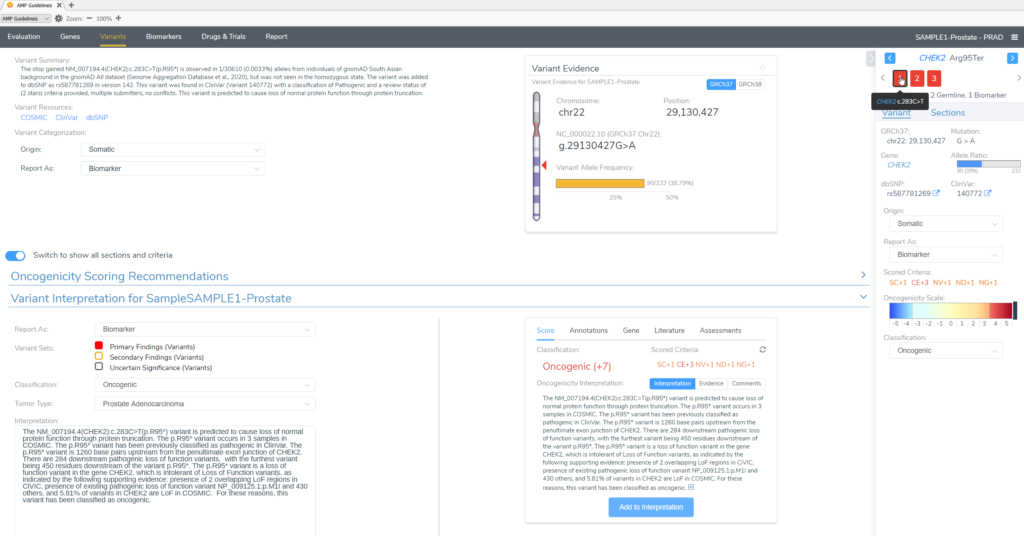
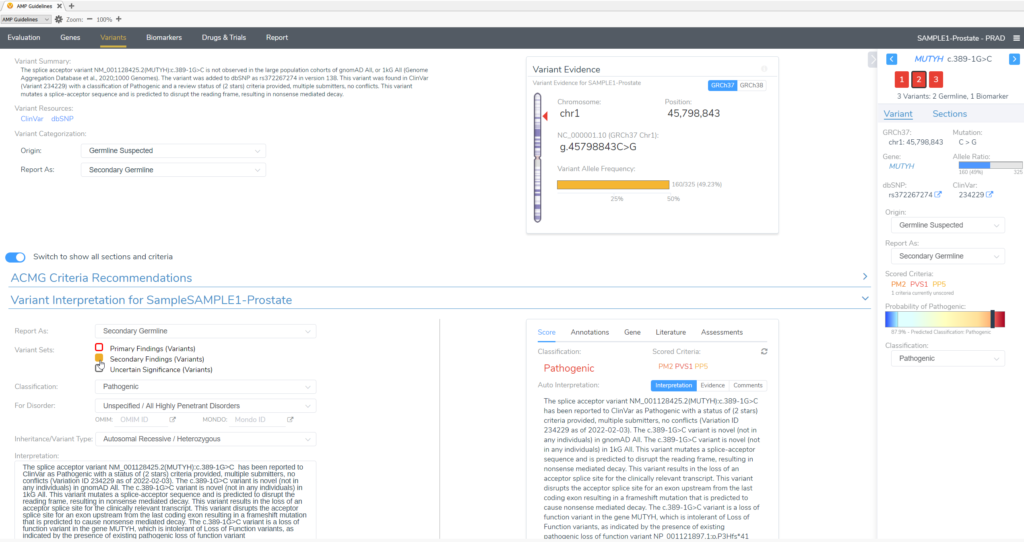
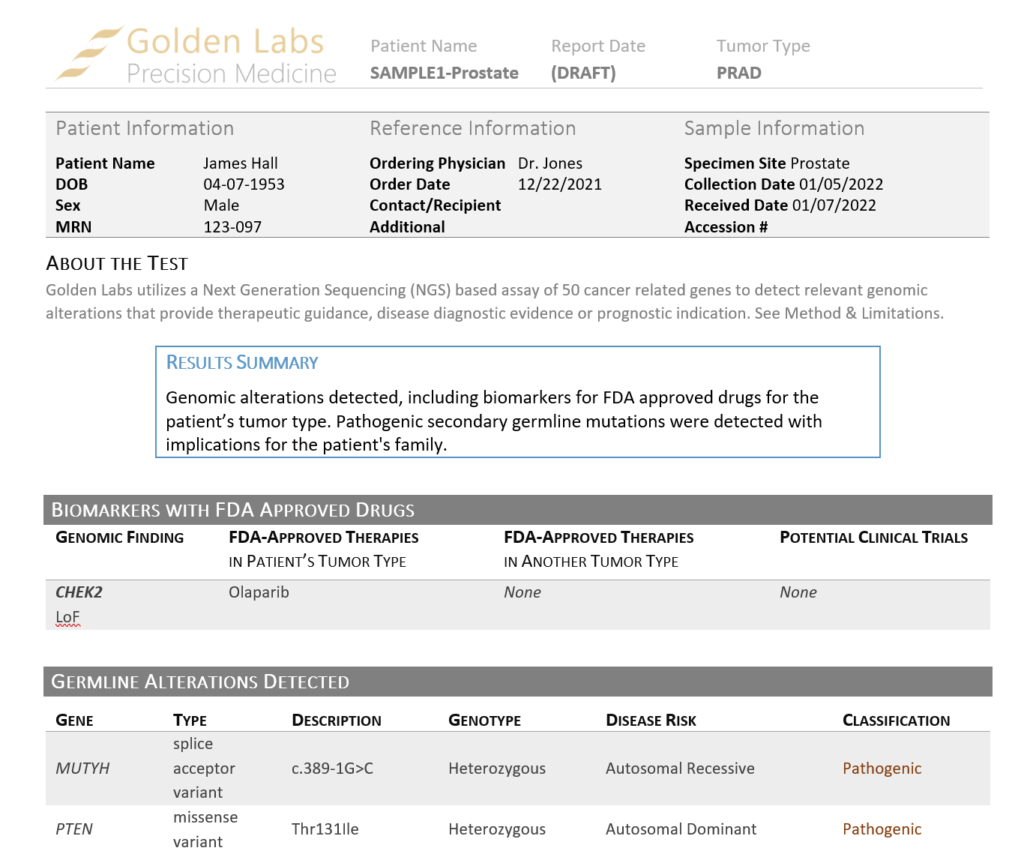
In this example of a prostate adenocarcinoma, we identified a somatic variant in the gene CHEK2 and two secondary germline variants in the genes PTEN and MUYTH based on our filtering strategy in VarSeq (Figure 3). Below the Filter Variants list, VSClinical gives us the option to select the origin of the mutations we are importing for analysis. Somatic was selected for the CHEK2 p.R95*, while Germline Suspected was selected for PTEN p.T131I and MUTYH c.389-1G>C. Already, it can be seen that these variants will be handled differently in VSClinical AMP, with the CHEK2 variant being classified as Oncogenic and being reported as a Biomarker according to our standard AMP guidelines designations, while the germline variants are being classified as Pathogenic and reported as secondary germline variants.
Our somatic variant will be analyzed according to the AMP guidelines, given an Oncogenicity score and brought in to the final report as a Primary Finding.
Conversely, our germline variants will be scored as Pathogenic, based on the ACMG guidelines for germline variants, and brought in to the final clinical report as Secondary Findings.
Our shipped somatic report templates are designed to accommodate secondary germline variants. Once the report is rendered using our Cancer Drugs and Trials Template V2, the somatic and secondary germline variants are reported in separate sections (Figure 6) and a note is made about each set of findings in the Results Summary.
The ability to easily render a somatic clinical report that includes secondary germline variants is an example of how VSClinical accounts for the wide range of clinical reporting needs. To learn more about clinical reporting in VSClinical, check out our other blogs and webcasts or reach out to us at [email protected].