With the release of VarSeq 2.2.4 just around the corner, I want to detail some new 2.2.4 features that will enhance somatic variant annotation and fusion analysis within VSClinical. A webcast back in July showed some of these updates in action, so if you are looking for some more content on this topic, I would highly recommend checking out the webcast as well!
Let’s start things off by mentioning two annotation sources currently available in VarSeq 2.2.3 but were recently updated and released, COSMIC 94 and Golden Helix CancerKB. Golden Helix has been curating COSMIC annotation sources since COSMIC 88, all available in VarSeq. The COSMIC annotation is broken up into 5 separate sources: Fusions, Hallmarks, Transcript Counts, Cancer Gene Census, and Mutations. These are now available with the most up-to-date data from COSMIC 94 and can be used to annotate VarSeq projects and in VSClinical. In the COSMIC 94 update, the added mutations’ general focus was on rare lung and pancreatic cancers. In addition, 12 genes that are known to be involved in apoptosis pathways were the main focus for this release. The cancer hallmarks annotation was updated to include functional information on 9 Cancer Gene Census Tier I genes causally associated with cancer. Ultimately, this information provides an improved understanding of how these genes contribute to tumor development.
I plan to spend more time going into the updates to Golden Helix CancerKB in a future blog, but I want to briefly mention it now as it ties into the changes made to analyze gene fusions in VSClinical AMP. Golden Helix CancerKB is a Golden Helix curated track wherein a panel of expert curators who have strong backgrounds in writing clinical somatic variant interpretations gather information from several sources to find the most up-to-date information to describe a gene’s function, impact, and alteration frequency in different cancer types. Interpretations are also written about the most prominent biomarkers in these genes. They are described in terms of their role in cancer, how common the mutation is for a given tumor type, and if the biomarker has any diagnostic or prognostic implications. And of course, on a therapeutic level, drug sensitivity or resistance information for the biomarkers are also investigated and captured into interpretations. An updated version of Golden Helix CancerKB was released in August 2021 and includes over 100 new interpretations and updates to over 180 existing interpretations with Tier Level I clinical evidence.
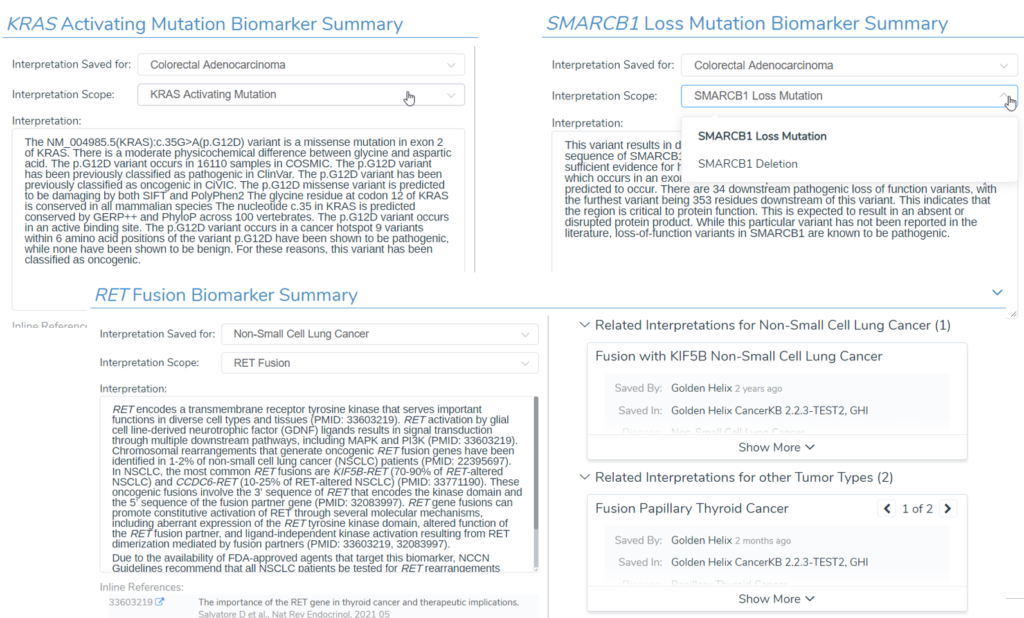
Part of this update to Golden Helix CancerKB included restructuring how interpretations were written and associated with specific biomarkers. An example of this can be explained in the context of gene fusions. Previously, Golden Helix CancerKB interpretations were written for a particular fusion pair, like KIF5B-RET in non-small cell cancer (NSCLC). However, RET fusions in NSCLC are not limited to fusions with only KIF5B. Having this in mind, the scopes for some Golden Helix CancerKB biomarker interpretations in some cases have been broadened, in this case to RET fusions in NSCLC in general. So to accommodate some of these more broadly scoped interpretations, updates were also made to VSClinical AMP to accommodate the broadened interpretation scopes.
In VSClinical AMP, the new biomarker scopes that can be selected for adding biomarker interpretations are Loss Mutation (complete gene deletion), Activating Mutation (any mutation resulting in proto-oncogene activation), and single gene Fusions. The new broadened Golden Helix CancerKB interpretations and new biomarker scopes in VSClinical AMP will be available in VarSeq 2.2.4.
Finally, to wrap up the discussion, I want to mention that we will be updating our Cancer Ontologies for VSClinical AMP. Research is constantly being done to better characterize cancers; for example, there has been a push to characterize hematological cancers by the WHO and other groups in recent years. In lieu of these efforts, we have updated the list of available cancer ontologies to ensure tumor type-specific interpretations can be recorded. In some cases, a mutation has implications across multiple tumors, such as all solid tumors or hematological cancers. To capture these use cases, we have also added the ability to create biomarker interpretations for solid tumors and hematological tumors. These ontology updates are also reflected in Golden Helix CancerKB interpretations, making filling in these interpretations much easier.
There is a lot to look forward to in the upcoming VarSeq 2.2.4 release, so stay tuned for more exciting content!