Clinical labs often maintain gene panels, which are lists of genes with evidence of disease association. These panels are used to prioritize variants and limit interpretations to a predefined set of test-specific genes. In general, gene panels should be stored independently of any specific project or interpretation, as it is common for an individual gene panel to be generally applicable to many different samples. Additionally, multiple gene panels are often relevant to a given patient or sample. To meet the needs of clinical labs, several well-respected organizations have developed standard lists of genes with clear evidence of disease association.
PanelApp
One such organization is Genomics England, which has developed PanelApp, a publicly available knowledge base of gene panels related to human disorders. PanelApp contains gene lists for a wide variety of disorders and each panel is versioned and validated by experts in accordance with a strict curation process. Each gene is classified into one of three status categories based on the level of available evidence:
- Green: These genes are considered diagnostic-grade and require evidence from three or more unrelated families or from two families where there is strong additional functional data.
- Amber: Genes with this status are associated with borderline evidence.
- Red: These genes are associated with a low level of evidence.
These panels are an excellent source for providing evidence supporting diagnostic links between genes and specific disorders.
ACMG Secondary Findings
Another useful gene panel resource is the ACMG Secondary Findings gene list. The American College of Medical Genetics and Genomics has published recommendations for reporting incidental findings in exome and genome sequencing data along with a corresponding list of genes, with the latest update to this list published earlier this year. These recommendations state that laboratories should report all occurrences of certain mutation types occurring in the listed genes as incidental findings. Since this list is updated periodically, labs must maintain an up-to-date list of these genes if they wish to follow these recommendations.
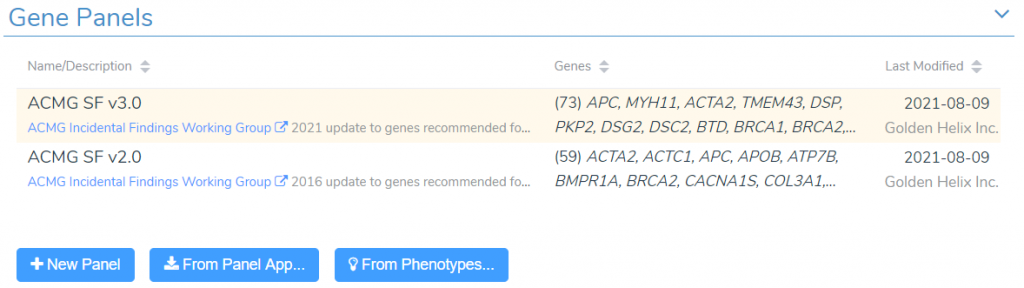
Manage Gene Lists in VarSeq
We are happy to announce that the upcoming VarSeq release will support the creation and management of reusable gene lists. This feature will include a customizable gene panel library containing the newest versions of the ACMG Secondary Findings gene lists. New panels can be easily added to this library using the gene lists supplied by PanelApp or through gene-phenotype associations in the HPO and MONDO ontologies. These panels can be easily integrated into your VarSeq workflows through new filtering functionality and new algorithms for matching variants and CNVs that overlap genes in your panels.
To manage our gene panels in VarSeq, we sill start by selecting Tools -> Manage Gene Panels from the menu bar. This will launch the gene panel management interface. At the top of this window, we can see the current list of panels in our gene panel library. By default, this list will include multiple versions of the ACMG Secondary Findings panels.
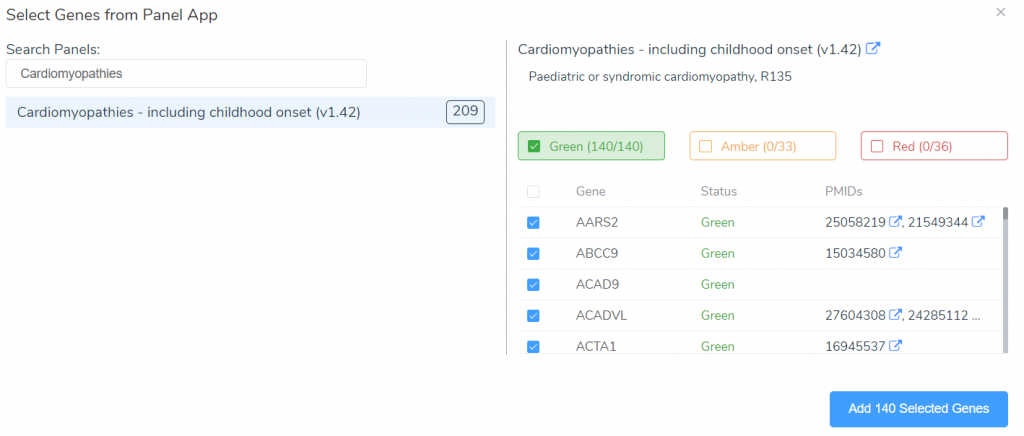
We can look at the details associated with any panel in our library by clicking on the panel name in the list. To create a new panel from scratch, we can click on the New Panel button. However, we will often want to create panels targeting specific disorders or phenotypes. When dealing with disorders that have well-established gene associations, we will most likely want to use the From PanelApp tool.
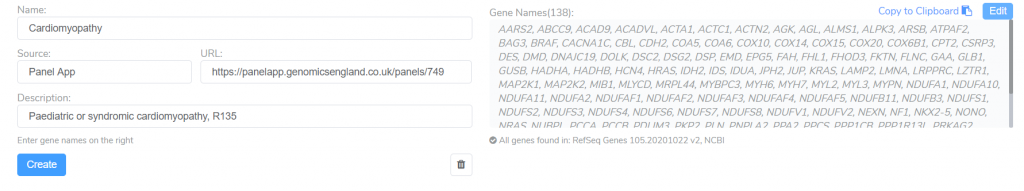
This tool can be used to search the PanelApp knowledge base for existing panels related to a specific disorder. These panels are an excellent starting point for creating your own gene panels targeting a given disorder. After adding the genes associated with this disorder in PanelApp, we can customize the panel by editing the name, URL, description, or by adding and removing genes.
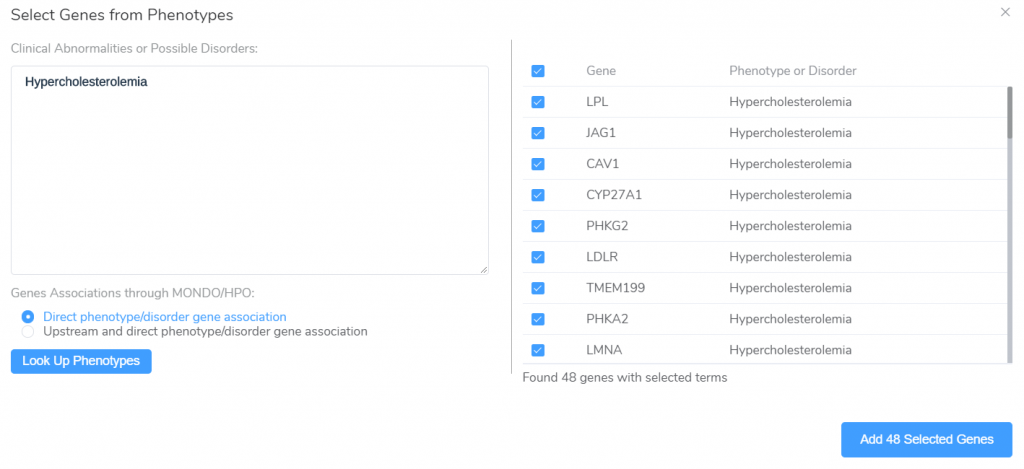
VarSeq also supports the creation of gene panels based on the gene-phenotype relationships in the HPO and MONDO ontologies. Clicking the From Phenotypes button will launch the Genes from Phenotypes panel creation window.
From this window, we can enter a list of HPO and MONDO phenotype/disorder terms and lookup genes associated with these terms, either directly or through association with more general upstream nodes in the ontology. The associated genes can then be used as a starting point for the creation of phenotype-specific gene panels.
To see this in action, head over to our on-demand webcast “Creating & Managing Reusable Gene Lists with VSClinical”. Or, you can request a personal demo with our team by emailing [email protected]!
Using Gene Panels to Prioritize Variants
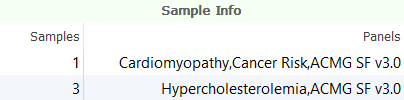
There are several tools for prioritizing variants based using gene panels in VarSeq. Gene panels can be associated with individual samples through the Samples Table by specifying a Panels field containing a list of gene panel names.
These sample-level gene panels can be leveraged to prioritize variants using the Match Panels (Per Sample) Algorithm. This algorithm determines matches between the gene annotations of each variant and the list of genes in the panels associated with each sample. By filtering on this annotation we can eliminate variants that do not overlap any genes in the gene panels associated with each sample.
Another useful algorithm for gene panel annotation is the project-level Match Genes and Panels algorithm. This algorithm identifies matches between variant gene annotations and a single user-selected gene panel.
Rather than utilizing the per-sample panel names specified in the sample table, this algorithm requires the user to select a single panel name from the list of panels in the gene panel library. Alternatively, the user may instead specify a list of gene names to match. This algorithm can be used to filter down to a set of variants associated with genes in a single specific panel or gene list.
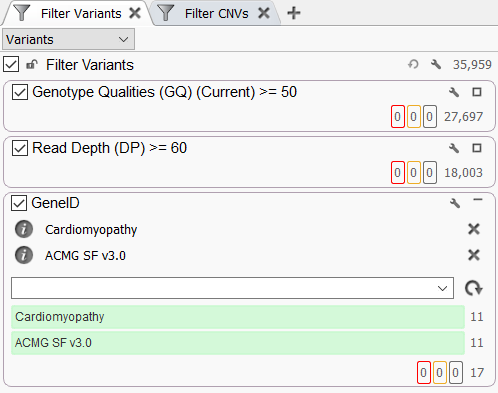
Variants can also be prioritized using the Gene Panel Filter in the VarSeq filter chain. This feature is particularly useful, as it allows variant filtering based on gene panels without requiring any additional algorithms. To add this filter, right-click inside the filter chain and select Add Gene Panel Filter. After selecting a Gene ID field, you will be presented with a new filter containing a drop-down menu that can be used to select one or more gene panels from your library.
This filter will exclude any variants that do not overlap genes in at least one of the specified gene panels. This can be a useful tool for quickly determining which gene panels contain clinically relevant variants.
Gene Panels in VSClinical
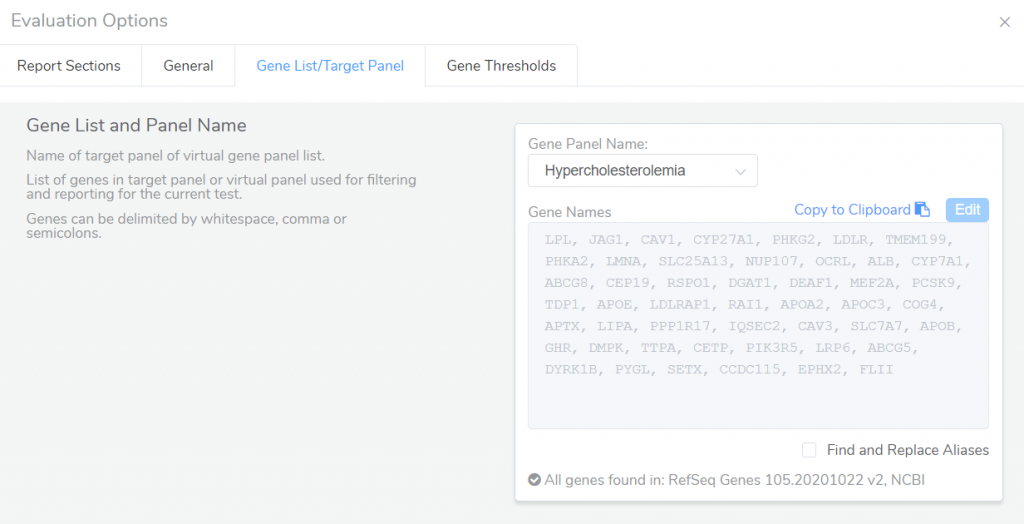
Gene panels can also be used in VSClinical to specify which genes are included in the final clinical report. Through the Evaluation Options in VSClinical, you can specify a specific gene panel containing the list of genes to be included in the report for the current test.
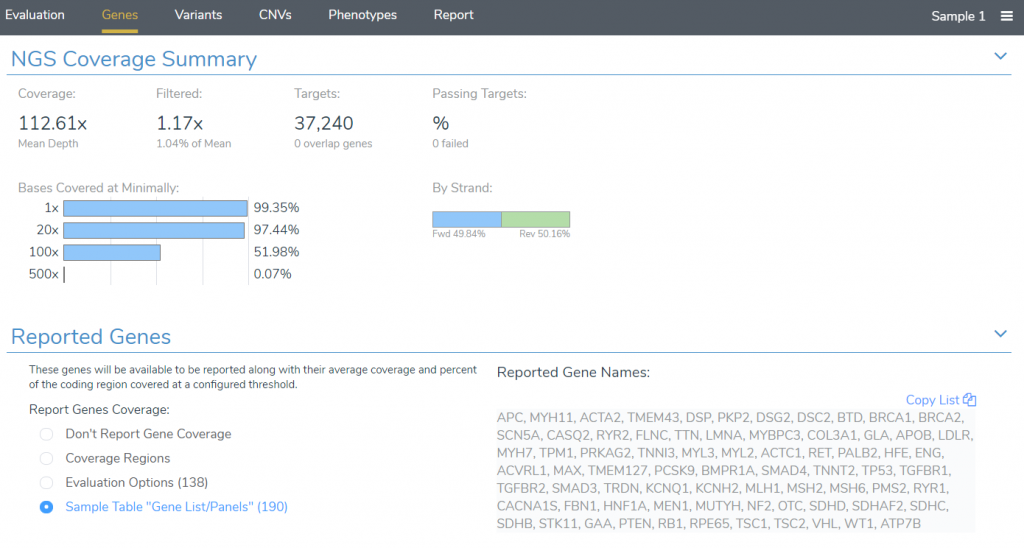
After starting an evaluation, reported genes can be specified in the Genes tab. By default, all genes associated with the panels listed in the samples table will be included in the report, but this default behavior can be changed to instead report genes included in the gene panel specified in the Evaluation Options.
Conclusion
In this blog post, we discussed the upcoming Managed Gene List feature of VarSeq and VSClinical, which enables the definition and re-use of gene lists outside of individual project templates. We are excited to announce the upcoming release of these new features which will become available later this year. If there are any questions about the content of this blog post, feel free to reach us at [email protected]!