In addition to Golden Helix providing easy-to-use genomic software, we also provide value to our users by automating the curation of public databases used in our tools. These annotation sources serve multiple purposes not only leveraging key fields in the filter chain but also automatically supplying evidence for the ACMG and AMP classifications reviewed in VSClinical. Supporting the curation of these databases is ongoing with the development of our products and every now and again we get some newcomers to the list. Here are some example tracks that are new or have had some recent updates:
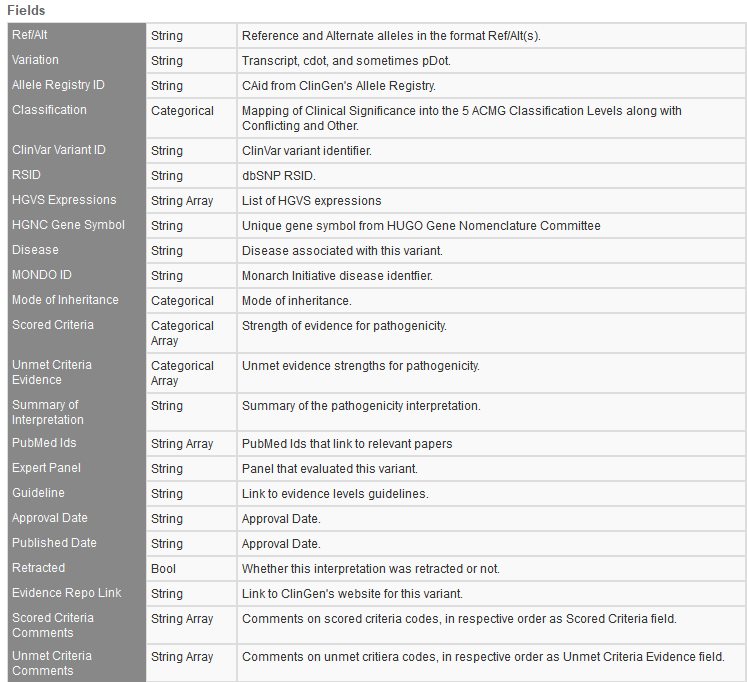
ClinGen Expert Curated Interpretation of Variants
ClinGen ECIV is funded by the National Institutes of Health (NIH) to provide an authoritative resource that defines the clinical relevance of genes and variants for use in precision medicine and research. This track is refined in presenting not only basic variant level format fields and notation but supplies the user with a full overview of a variant ACMG based classification plus associated disorders and details on the expert panel that reviewed the variant. You can see a breakdown of the fields in Figure 1.
The ClinGen track supplies a major level of convenience to the user when evaluating variant classifications since most of the hard work of classification is already done and presented from the database.
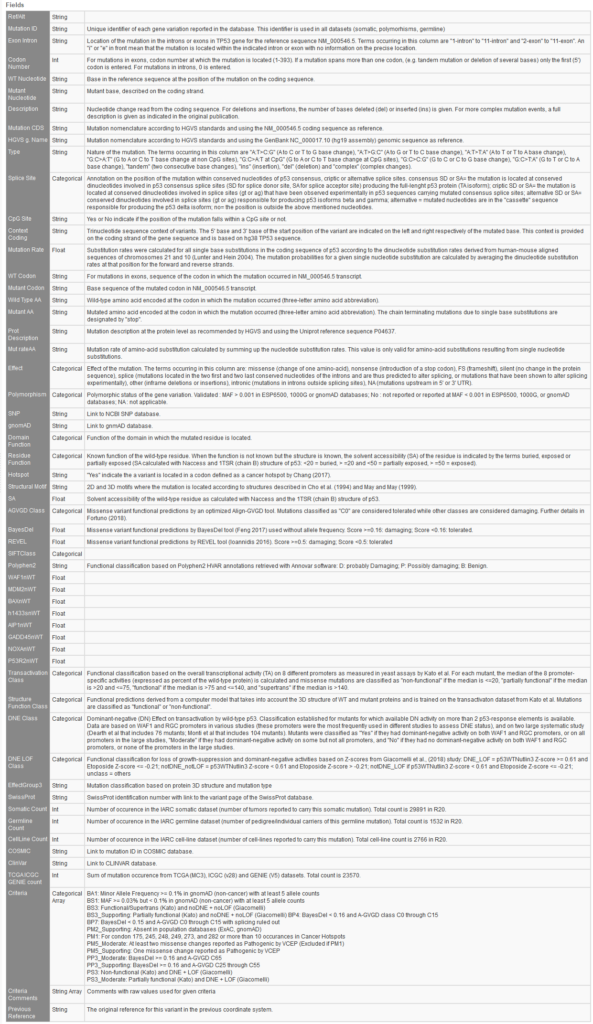
International Agency for Research on Cancer (IARC) TP53 ACMG R20
Likewise, the new ACMG TP53 database also compiles various data relevant to TP53 gene variations relevant to cancer. This database is extensive in providing the user ample data on variant notation, impact, functional prediction, domain function, number of submissions, and relevant ACMG criteria.
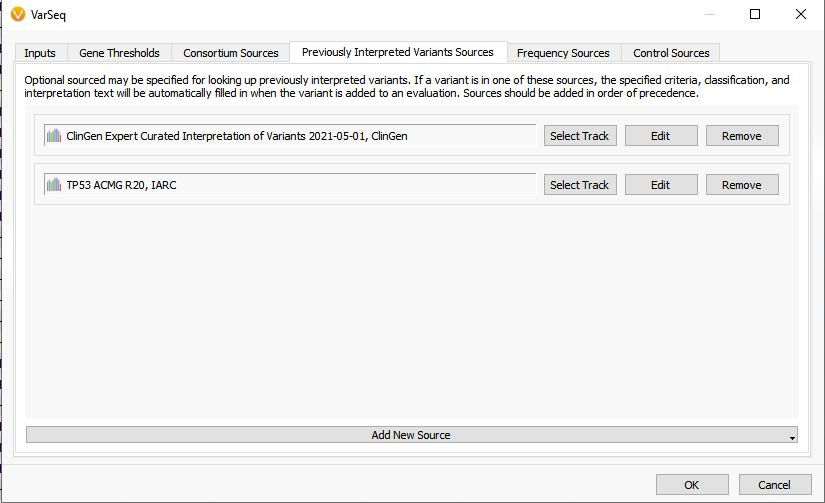
Both the TP53 and ClinGen sources now are factored into the ACMG auto-classification built directly into the VarSeq software. Figure 3 shows the option to adjust the ACMG classifier to now include the ClinGen and TP53 sources as default inputs for the Previously Interpreted Variant Sources. Overall, this will help automatically adjust for criteria specific to the TP53 gene and evaluated variants stored in ClinGen. To see a demonstration of this value, please watch this recent webcast.
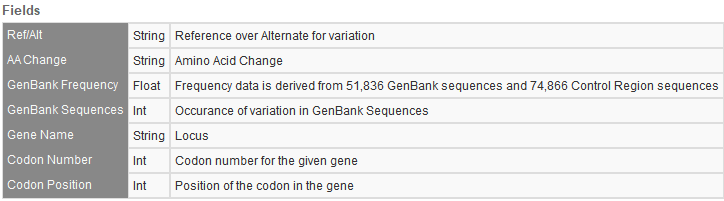
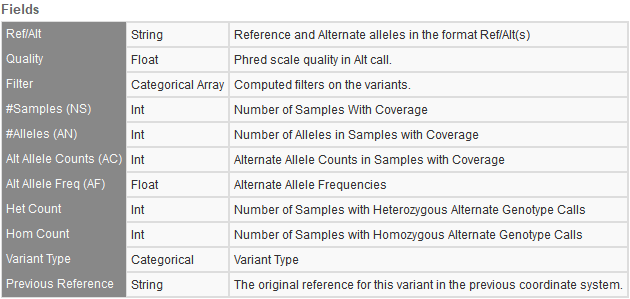
NCBI dbSNP 155
This commonly used variant database is massive and comprised of single nucleotide variants, insertions and deletions, short tandem repeats, and microsatellites. This upgrade has now bumped the number of variants up from the last versions 717,580,549 variants to 1,246,001,635 variants. Figure 4 shows the fields provided from the dbSNP database.
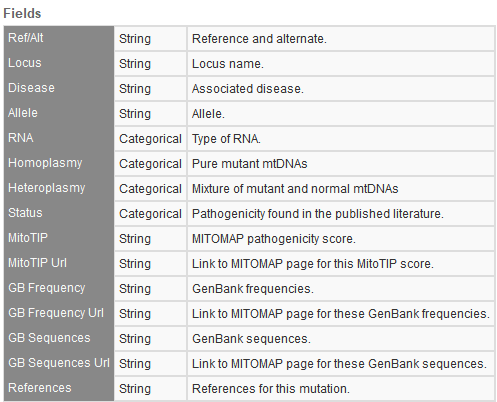
MITOMAP Coding and Control Region Mutations, Polymorphisms, rRNA/tRNA Mutations
It is becoming more and more frequent that our users seek to understand the impact of mitochondrial variants. This has driven our support to provide the latest MITOMAP databases to open the door to these evaluations. You will see snapshots of these fields in Figures 5a-c below.
TOPMed Variant Frequencies Freeze8
TOPMed or the Trans-Omics for Precision Medicine as submitted to dbSNP
Across more than 80 different studies and over 158K samples, this track supplies a massive amount of variants to assist with variant allele frequency filtering. With the objective being to isolate clinically relevant variants for rare disorders, utilizing alternate allele frequencies in the filter process provides a huge advantage at eliminating uninteresting common variants. This track used in conjunction with other massive databases such as 1kg Phase 3 or gnomAD exomes/genomes allows for powerful filtering when setting the typical clinical allele frequency thresholds to 1% or less in the population.
If you wish to apply these databases to your workflow, you’ll find them in the VarSeq Manage Data Source Library under Public Annotations. To find out more on how to best utilize VSClinical and the VarSeq Data Source Library, please email us at [email protected].