One of the many tricks of encoding so much functionality into so little space in eukaryotic genomes is the ability to produce multiple distinct mRNAs (transcripts) from a single gene. While one transcript is often the dominant one for a given tissue or cell type, there are, of course, exceptions in the messy reality of biology. It doesn’t take many times through a variant interpretation workflow before you gain an appreciation for selecting gene transcripts. Yet when describing a variant on a genetic report, a choice of transcript must be made to provide an HGVS coding and protein description of the mutation. In some cases, the choice of transcript may change the variant from being described as exonic to intronic or from a loss-of-function pathogenic mutation to a non-coding benign length polymorphism. Having accurate and complete transcript models and picking the most biologically and clinically relevant transcript is thus an important choice when completing rare variant interpretation for genetic testing.
Annotating Transcripts First, Ask Questions Later
While a single transcript choice must be made eventually, variant analysis should not start with a narrowed focus on a single transcript. The VarSeq gene annotation algorithm performs a one-to-many annotation of a variant against every overlapping transcript. By default, the annotated transcripts are filtered to mRNA, and non-coding RNAs as the complete set of RefSeq genes transcripts include many experimentally predicted transcripts (with an “XM_” prefix) that should generally be ignored.
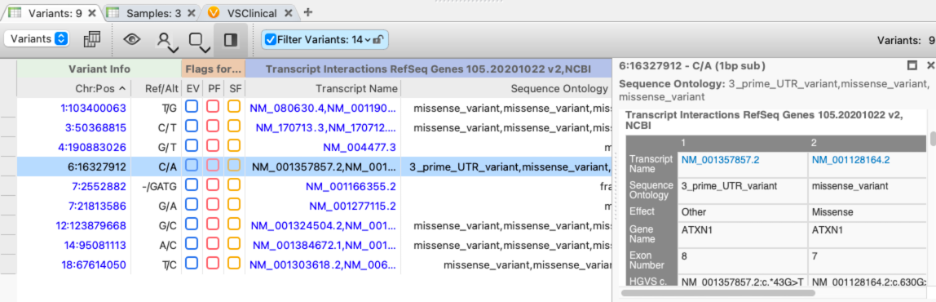
To handle the presence of multiple transcripts, the VarSeq gene annotation algorithms produce three column groups:
- Summary fields that combine the per-transcript annotations
- Transcript Interactions that include the per-transcript annotations
- Aux Fields that pass through the transcript-level additional auxiliary fields from the annotation source
To aggregate the per-transcript annotations into a single value that can be used for filtering or exporting the variant table in a meaningful way, there are two strategies and corresponding fields:
- Combined: For fields like Sequence Ontology and Gene Region, this takes the “worst” annotation result from all transcripts. Useful for conservative filtering.
- Clinically Relevant: Pass through a single transcript’s annotations. This depends on the choice of a “clinically relevant” transcript.
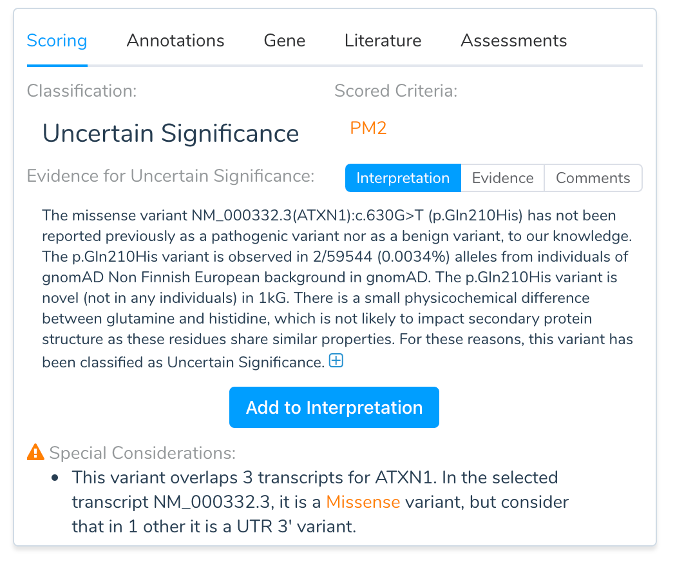
While VSClinical adds variants to an evaluation based on the Clinically Relevant transcript, it will also warn when the annotation differs in effect on other transcripts.
VSClinical supports switching the analysis of the variant to another transcript at any time. When switching transcripts, a variant is updated to reflect the per-transcript annotations, sequence ontology, in-silico functional predictions, canonical and novel spicing effects, and ultimately the recommended criteria following the ACMG guidelines.
The Clinically Relevant Transcript
So how does VarSeq select the “Clinically Relevant” transcript from the many a variant may overlap? Well, because there is no pre-defined “best” transcript in annotation sources like RefSeq for every gene in the genome, a heuristic must be used. We have put a lot of effort into building this heuristic to match the leading variant annotation sources in the industry, specifically ClinVar. Over time, we have updated it to include more annotations and clinical databases such as ClinVar, LRG, and, most recently, MANE. In our previous blog post What’s in a Name: The Intricacies of Identifying Variants we outline this heuristic and how it closely matches ClinVar’s default transcript selection. Now updated to prefer MANE “Select” transcripts over LRG transcripts, the heuristic is as follows:
- Prefer a transcript that is a MANE “Select” transcript
- Prefer a transcript that has an LRG identifier
- Prefer a transcript that has correctly encoded start and stop codons over “incomplete” transcripts
- Prefer a transcript that is protein coding over one that is non-coding
- Prefer transcripts with longer coding sequences
- If all else is identical, select the first in lexigraphic order
As a joint effort of the NCBI gene annotation and the Ensembl gene annotation folks, the Matched Annotation from NCBI and EMBL-EBI (MANE) transcript set aims to define a single representative transcript per gene that is genomically identical between both annotation team’s databases and “is well-supported by experimental data and represents the biology of the gene.”
In practice, for the roughly 18,000 genes in the MANE 0.93 release, a “Select” transcript based on computation tools such as per-tissue expression will be selected as the Clinically Relevant transcript. That is unless that choice is overridden by a saved user or system gene preference.
The Gene Preferences Override
While the described clinically relevant transcript heuristic provides a baseline solution, it does not consider the clinical community’s momentum on choosing one transcript over another for clinically reporting and publishing. For this reason, we ship a system “Gene Preferences” file that specifies a specific transcript for certain common clinical genes. See Using Gene Preferences in VarSeq and VSClinical to learn more about what Gene Preferences cover and how the user saved preferences always take precedence and ultimately allow any of these preferences to be set explicitly by the user and lab.
There is generally a dominant transcript in use for genes that have been heavily used in clinical genomics. We have found that ClinVar submissions of variant assessments to be a good source of transcript preferences of clinical labs. These submissions include a transcript name as part of the HGVS for a variant. For a given gene, we thus count the frequency of each transcript (ignoring the transcript “version”) in the current set of ClinVar variant submissions. Note this data is available to you as the “ClinVar Assessments Counts” field in the latest monthly curation of the “ClinVar Transcript Counts” track. It can be a great resource for plotting and inspecting genes and is also displayed in the VSClinical transcript selection dialog.
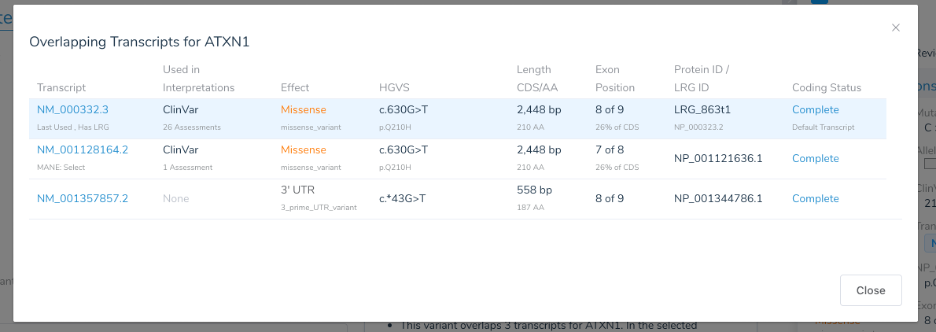
For commonly tested genes, these submission counts can be in the thousands. On the other hand, less commonly analyzed genes may have just a few submissions. After doing analysis and case studies of commonly tested genes, we developed the following heuristic for adding a transcript preference to our system gene preferences file (thus overriding the default choice, often provided by a MANE transcript):
- If a transcript has the most ClinVar submission references
- … and it has a submission count greater than 10
- … and it is not the default transcript choice
- … and the default transcript choice does not have a submission count greater than half of the most submitted transcript
- Then: add the most submitted transcript as a manual transcript selection in the system gene preferences
Here are a few examples to enrich this logic with some real-world context:
BRAF: In recent gene transcript annotations, BRAF now has 13 transcripts. One of those (NM_001374258) has been chosen by MANE as a MANE “Select” transcript. Yet over the many years of clinically testing of BRAF, NM_004333 has been the most reported and published (most likely due to being the first annotated transcript). The NM_004333 transcript has 563 ClinVar submissions and, through this heuristic, is saved in the VarSeq system Gene Preferences as the default Clinically Relevant transcript, overriding the default MANE select transcript.
CERKL: Sometimes, the clinical testing community preference is not clear cut. In gene CERKL, the default choice of transcript is NM_201548. Looking at the ClinVar submission count data, a different transcript NM_001030311 ranks highest with 172 submission counts. But NM_201548 has a considerable 97 submissions, more than half of the first ranked transcript. We feel the lead here in submission counts is not strong enough to out-weight the MANE “Select” transcript choice. So, this gene does not get a transcript override, and NM_201548 remains the Clinically Relevant transcript.
Genes Are Not Static
I wouldn’t have expected to be writing this blog post a few years ago, as it seemed for a while that our model of genes on the human genome was high quality and didn’t need much updating. But in the last couple of years, we have seen massive updates to transcript models, continuous updates to gene names by HUGO, and efforts of new groups like MANE to make transcript choices based on empirical evidence. We have put in the work to keep VarSeq current with these changes while still maintaining the ability of labs to lock down and preserve the versions of critical annotation sources they use in their day-to-day analysis. For this reason, we plan to regularly ship the latest gene tracks to our annotation servers. Still, we very rarely make further changes to the bundled VarSeq gene tracks and system gene preferences.
VarSeq provides an excellent up-to-date platform to annotate and interpret clinical variants with its comprehensive annotations and carefully made default choices. As you can see in this post, much work happens behind the scenes to make your life following the ACMG or AMP guidelines for variant interpretation easier. Have an interesting gene or variant with tricky transcript or gene annotation issues? Reach out to [email protected]; we would love to see it!