Next-generation sequencing generates an immense amount of data which is then subject to a multi-step process to establish a validated bioinformatic pipeline. From processing raw sequence data to the detection of genetic mutations, establishing a validated and consistent bioinformatic pipeline makes a huge difference in the quality of patient care and accuracy of results. In this blog, we are focusing on how you can lock in annotation source versions as a part of your workflow. The Golden Helix software suite automates the NGS pipeline so much so that users can start with FASTQ files and eventually generate clinical reports in almost one step! Importantly, through the various automation steps, a key point is that consistency is always maintained. We have many other blogs, ebooks, and webcasts that discuss these automation tools in greater detail, however, in this blog I will discuss two ways that VarSeq can be used to ensure consistency when it comes to variant annotation and how this step can be automated for VarSeq projects.
A feature that has always been included with VarSeq is that users can create customized project templates which can be utilized as new data is acquired. This is a great way to automate project creation as you do not have to start from scratch creating projects. Using project templates also increases consistency. You can imagine a template that includes specific annotations which will automatically load, with a tailored filtering logic that will always be applied for any sample that is imported. But a finer detail I want to discuss is that custom project templates have the ability to lock the versions of annotation sources that will be used for all projects built from that template. Annotation sources are always evolving and being updated, which can be a challenge when it comes to validating your bioinformatic pipeline. However, being able to lock the versions of the annotation sources that will be used in all projects is one way to mitigate this issue. Figure 1 shows an example of various annotation sources and their versions which are locked into the project template.

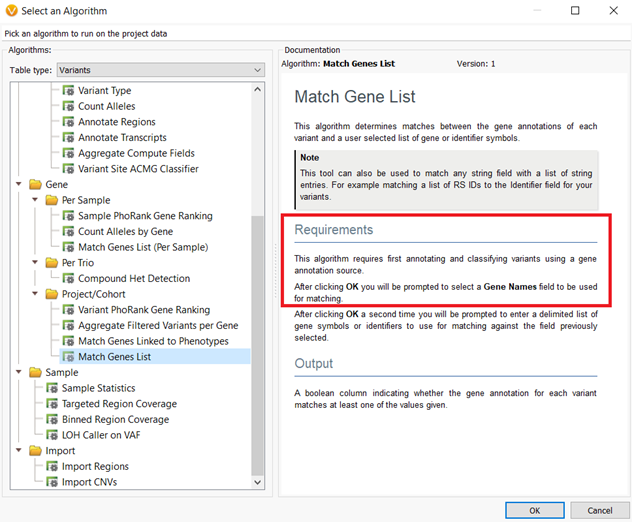
The annotation sources that are locked into project templates are those that will be used not only for variant annotation and filtering but also will be the source versions used for algorithm computation. An example of this concept is that the Match Genes List algorithm requires the annotation source RefSeq Genes for gene name input. Whichever version of RefSeq Genes is saved to the project template will be used for the Match Genes List algorithms’ computation.
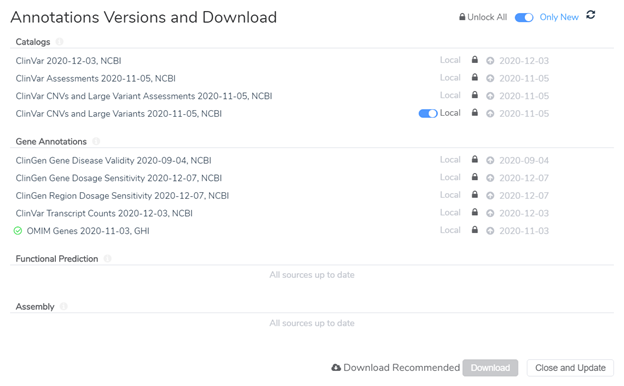
Locking annotation source versions to a project template is only step 1. Before actually saving the project as a template, we will need to make sure that the annotation sources are locked at the evaluation level as well. This second step applies to users who evaluate variants within VarSeq Clinical (VSClinical) according to the AMP or ACMG Guidelines. VSClinical requires additional annotation sources to complete variant analysis and generate a clinical report. After opening up VSClinical, users are prompted to download required sources and can lock these source versions so they will be used in all future VSClinical evaluations. Figure 3 shows an example of the source version dialogue within VSClinical. You will notice that I have locked different versions of the annotation sources that I would like to be used in all evaluations as noted by the dates on the right side of the image.
Once the annotation source versions are locked on both the project and evaluation levels, the project template can be saved and those annotation sources will be locked for all future projects that are made with that project template, thus ensuring consistent variant annotation across all projects and evaluations!
Choosing your locking preference on an annotation source level was not always a feature available for VarSeq or VSClinical. Historically, annotation sources that are required for VSClinical would utilize the latest annotation source that was downloaded and available. VarSeq project templates have previously been version-locked for all annotation sources. The ability to lock down specific annotation sources but allow others to update to the latest available was introduced in VarSeq 2.2.2. This allows maximum control for validating clinical pipelines while not missing any useful annotations in frequently updated sources like ClinVar.
If you have questions about locking annotation source versions on a project level or at an evaluation level, the field application scientist team is always available via email at [email protected] and we would be happy to schedule a training call to help! Also, if you are curious about other VarSeq tools and functionality to automate your clinical NGS workflow, we can also discuss these options and help integrate them into your existing workflows. Feel free to also check out some of our other blogs that contain important, useful news and updates for the next-gen sequencing community.