Type 2 Diabetes, Rheumatoid Arthritis, Obesity, Chrohn’s Diseases and Coronary Heart Disease are examples of common, chronic diseases that have a significant genetic component.
It should be no surprise that these diseases have been the target of much genetic research.
Yet over the past decade, the tools of our research efforts have failed to unravel the complete biological architecture of these diseases.
The most widely employed tool in the past 7 years for this research has been Genome Wide Association Studies (GWAS).
Now, Next Generation Sequencing is gaining traction as another tool to investigate the link between genetics and common diseases.
So let’s take a moment to evaluate the past 7 years of effort. What have we gained, and what lessons can we take into future research.
GWAS Studies Provide Correlation, not Causation
With a search space as big as the human genome (3 billion base pairs), you can’t start investigating the biology of a disease without knowing where to start looking.
This is what we have been doing for the last decade. Creating an association map between diseases and genetic regions (some of which may be in or near genes).
So we needed two things to build this map.
- Genetic markers from a wide range of genomic regions
- A tool to correlate those to diseases
The correlation between a marker and a disease is just a hypothesis (the null hypothesis being the marker and the disease are not correlated).
Aside from common confounding issues (such as signal-to-noise problems, multiple-testing, and variables other than disease status dominating the experiment), using a statistical hypothesis test such as the Pearson’s chi-squared test can give us the probability that that there is no correlation between a given genetic marker and our disease status.
In the best case, we find some markers that have a very low-probability that their correlation to the disease status is by chance alone; we then build up, over time, a list of associated SNPs (common variants) to the disease.
The trick is, you are not running just one test in a Genome Wide Association Studies (GWAS), you are running hundreds of thousands of tests.
To have enough statistical power such that you can have confidence that your association is not by chance alone, your experiment needs to have thousands if not tens of thousands of samples. Each one of those samples must be genotyped using a SNP array, which historically has cost hundreds of dollars per sample.
Were These GWAS Studies Worth $100 Million Dollars?
That doesn’t sound cheap, does it?!? These studies can cost millions of dollars and, by their nature, usually end up being executed by a consortiums that pool funding and man-power over many institutions and even countries.
Yet, by the estimates of the NHGRI, around 1,000 GWAS studies have been published between 2005 and the end of 2011. Around 600 diseases or traits now have mapping to the genome (you can just as easily put “head circumference” as your variable as well as “do you have Type-2 diabetes”).
Note some important limits:
- The results are valid only for the set of samples in the experiment, which hopefully represents the samples outside the experiment, but may not for all ethnicities or familial backgrounds.
- It is still possible the correlation discovered is not with the disease we are interested in, but is with some other variable that happens to align well with the disease variable.
- The markers we can test must be relatively common (generally showing up more than 1 out of 100 individuals).
- The implicated genetic markers don’t directly explain the genetic architecture of the trait in a causal biological sense.
- Because so many samples are used to gain statistical power, the marker association can be statistically significant while the marker still has a very small effect size (a good explanation here).
In fact, very few trait-to-genotype associations from GWAS studies have shown a large effect size (strength of the association).
This means, your genotype for any given GWAS SNP is not likely to dramatically change your risk for the studied disease.
Even taking the combined effect size of all associated SNPs for a given trait, the effect size is not as large as we would expect (given how much of that trait we think is due to genetics versus the environment).
This dilemma has been termed the “missing heritability” problem.
A representative example is Rheumatoid Arthritis, where we estimate about 60% of your lifetime risk of this condition is due to your genetics. Yet, a recent study done at the Broad Institute using thousand of GWAS SNPs shows we can only account for a little over a half of that heritable risk.
Companies like 23andme who very carefully examine every SNP association to make sure it meets their quality standard before incorporating them into their risk estimation models will account for even less.
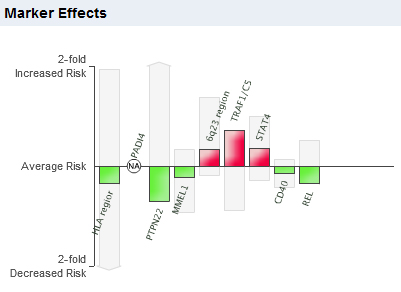
My 23andme risk prediction for Rheumatoid Arthritis estimates a 2.1% lifetime risk versus the 2.4% for the general population. The 8 markers shown above were implicated in GWAS studies and have relatively small effect size.
So has it been worth it to spend over $100 million dollars in research funding on these studies over the past seven years?
Yes.
But not because we discovered lots of actionable genetic markers. We haven’t.
And not because we have achieved a genetic understanding of common (and costly) diseases as we promised in our grants. We haven’t.
But science isn’t about delivering on a business plan.
Science is about discovery; breaking ground on venues of research that were previously entirely uncharted or unknown.
Already, follow-up studies are taking a deeper look at the genomic regions associated with certain traits.
Some of these studies are looking to close the gap of missing heritability by using Next-Generation Sequencing and new hypothesis about the biological architecture of common and chronic diseases.
With the expectation that genetics will play a large role in how clinical practice of medicine approaches preventative and personal care, there is an enormous amount of research left to make an individual’s genome actionable.
I’ll be watching closely.
I agree that ultimately, like most basic and applied research, it was worth spending $100M. The cumulative knowledge from spending this money will likely produce a larger return on investment for academic research and the biotech sector. However, in my opinion, GWAS were (and are) a waste of resources. Particularly because many (if not most) GWAS were poorly implemented, with the results being no different than a study in Phrenology. Association means very little without any investigation into causation. And very few GWAS have produced solid leads as to the cause of the association.
I wanted to supplement this post with a referral to the recent AJHG article I just read Five Years of GWAS Discovery.
They give many specific examples of things we gained from GWAS studies other than our low effect size associations, and come to a similar conclusion that the journey was worth it.
@Dave: I agree with you there there was even more blatant wast by those who didn’t properly set up their experiment design and simply focused on the more sexy aspects of a multi-million dollar project (namely, spending millions on genotyping thousands of people). But we have had plenty of posts on this blog about experimental design, so I thought it best to focus on the more controversial question about net value of our GWAS studies, even including those we tout as success stories.
Being critical of scientific research is all too easy when you know a little bit about the subject. If one really wants to be constructive then one should be supportive and encouraging.
The majority of GWAS have only looked for main effects which seems to me like a reasonable biological reason as to why the heritability accounted for is incomplete. For example, interactions with other SNPs and with the environment, not to mention dominance, recessivity, heterozygote effects, mosaicism, aneuploidy, pleiotropy etc are all established contributors to phenotypic diversity. See Manolio et al’s 2010 explainations for missing heritability http://www.ncbi.nlm.nih.gov/pubmed/19812666
GWAS was partially based upon the common disease/common variant hypothesis which is fairly loose. It seems to work for ApoE/Alzheimer’s but not much else. Twerlliger & Hiekkalinna’s 2006 ‘Utter refutation of HapMap (and GWAS) is an interesting read.
Then there’s Fisher and Visscher’s infinitisimal models….and rare genetic causes of common disorders, identified through exome sequencing and CNV arrays. Heritability estimates for common diseases are often very old and from small samples. Thus they may be incorrect. Common diseases clearly are not only genetically heterogeneous in terms of which loci cause disease but also in terms of the mode of inheritance – some RV, some CV, some in-between.
The GWAS data is easier to generate, store and analyse than NGS data. It has provided human population genetic data at a previously unpresidented scale, which can be analysed for many decades. Also, it has encouraged many small research groups to combine samples and form consortia. I’m not saying I’m ‘totally’ against whole-genome NGS though.
I once heard the question ‘How much money would be needed in order to find the genetic basis of common diseases?’ shortly followed by the popping of champagne corks in Illumunia offices. I hope that human biological research benefits all humans and not just those in the West.
Thanks for the comments. You definitely brought up some great examples of things the GWAS-era analysis projects have brought to the genetics community. For example, the need for more consortia level collaboration between groups to drive up sample size and distribute cost.
One thing I would beg to differ is on your comment about heritability estimates being based on old data with small sample sizes. The RA paper I referenced in Nature is basically the most up to date heritability estimate you could hope for, and is based on a huge mega-analysis of RA and other complex disease research.
Pingback: What can exomes tell us about the pathogenicity of complex disorders? | Our 2 SNPs…®
This is a great blog. Like human, why groups working in plants on GWAS not share their ideas behind their studies where most of the traits are quantitative. The studies conducted in plants for association mapping are successful or not, or trait-associated-regions are taken further for functional characterization, or post-GWAS application of TAS are important issues which can be discussed in details in such type of blogs. The comments from experts will be very much beneficial for those who just have started to work with GWA studies.