Clinical labs offer a unique and sophisticated product that is performed repeatedly with high standards of quality. VarSeq was developed to provide labs with the customization required for clinical genetic tests in a repeatable workflow. On top of this, VSClinical offers additional parameters and choices that can be made when designing the test workflow. In this blog series, we will deep-dive into these options and considerations at play when configuring your NGS testing process with VSClinical.
Your Lab’s Internal Interpretation Knowledgebase
The most important consideration when setting up your clinical workflow within VSClinical is configuring your internal knowledgebase. VSClinical saves every interpretation and classification of variants in a variant catalog deeply integrated with the rest of the ACMG workflow. Not only will this catalog be queried for each variant seen in any future samples tested, but it also constructs a landscape of interpreted variants across the genes being tested, providing supporting evidence in the proximity of newly evaluated variants. The variant catalog will show up in the table of clinically interpreted variants as well as the genomic plotting. These multiple use cases mean that as the number of users in the lab increases, the choice of catalog storage technologies becomes critical to the concurrent utilization of the catalog.
When configuring the VSClinical workflow for the first time, you will need to create a variant interpretation catalog in one of three types:
- File-Based: For individuals or small teams that infrequently perform variant interpretation at the same time, the SQLite file-based variant database on a network share can be a good starting point.
- Relational Database: VSClinical can store and query the variant catalog in a central database server and support multiple concurrent users. This is a great option for tech-savvy users that have an existing MySQL or Postgres relational database server in-house and are comfortable writing their own code to integrate this database into other lab systems.
- VSWarehouse: Backed by a relational database that also can scale to whole genome sized datasets, VSWarehouse can host the VSClinical interpretation catalog while also providing a web interface, an API and per-user permissions for catalog access. For most labs that want longitudinal storage of their genomic data and an interface to manage their lab’s interpretations over time for a number of users, this is the ideal solution.

VarSeq also supports exporting the variant interpretations from one catalog and loading them up into a new catalog. This allows a lab to start with a simple-to-setup file-based catalog and move to a VSWarehouse catalog as the number of active users increases or the need for catalog management and integration developments.

Re-using your VSClinical Settings
In addition to selecting the correct database type for the assessment catalog, the first time you run VSClinical, you can configure a few other useful options:
- Assay Name: When configured, this name will be saved in the assessment catalog with every interpretation.
- Input Record Sets: The way to select variants to evaluate using the ACMG guidelines, VarSeq supports creating one or more recordsets. Each recordset creates a column of colored checkboxes in the variant table, allowing for the easy selection of filtered and QC’d variants to evaluate.
- Report Record Sets: If using the VSReport module to create customized reports, classified variants can be placed into different sections of the report with these optional mappings.
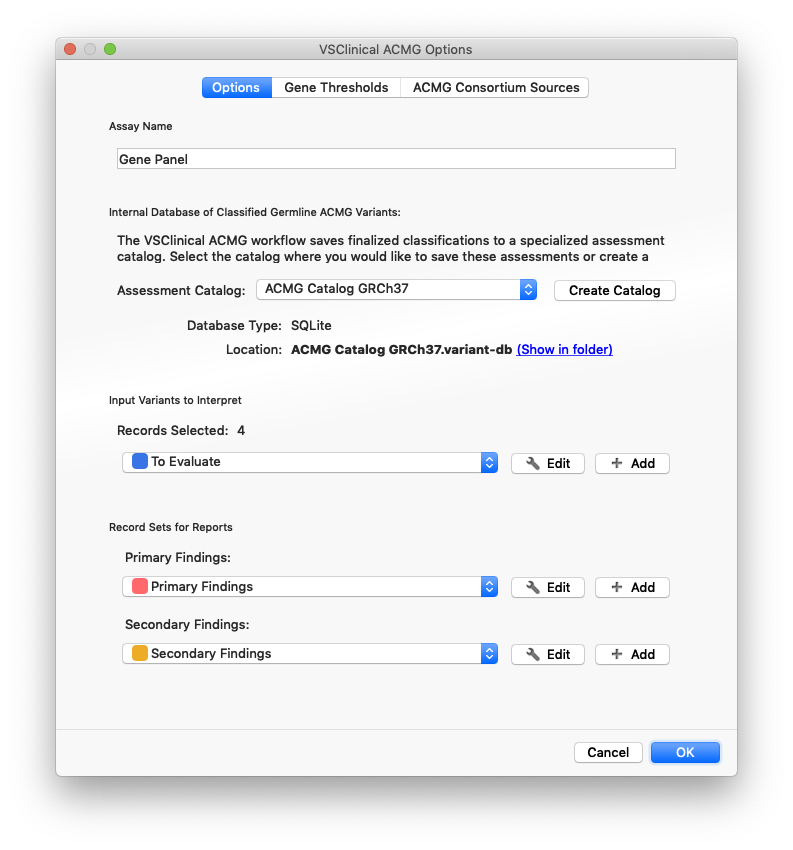
All of these settings are re-used when you turn your first project into a Project Template, allowing the exact same import, annotation, filtering, and VSClinical settings to be applied to the next batch of samples.
In the next part of this series, we dive into some of the advanced options for VSClinical that go beyond the basic configuration to tweak the way ACMG recommendations are made.